Abstract
Molecular tools have shown an added value in the diagnosis of infectious diseases, in particular for those caused by fastidious intracellular microorganisms, or in patients receiving antibiotics before sampling. If 16S rDNA amplification had been gradually implemented in microbiology laboratories, specific real-time polymerase chain reaction (PCR) would have permitted an increase in the sensitivity of molecular methods and a reduction of contamination. Herein, we report our experience in the diagnosis of infectious diseases over two years, during which 32,948 clinical samples from 18,056 patients were received from France and abroad. Among these samples, 81,476 PCRs were performed, of which 1,192 were positive. Molecular techniques detected intracellular microorganisms in 31.3 % of respiratory samples, 27.8 % of endocarditis samples and 51.9 % of adenitis samples. Excluding intracellular bacteria, 25 % of the positive samples in this series were sterile in culture. Conventional broad-range PCR permitted the identification of fastidious and anaerobic microorganisms, but specific real-time PCR showed a significant superiority in the diagnosis of osteoarticular infections, in particular for those caused by Kingella kingae and Staphylococcus aureus, and for endocarditis diagnosis, specifically when Streptococcus gallolyticus and Staphylococcus aureus were involved. The sensitivity of conventional broad-range PCR was 62.9 % concerning overall diagnoses for which both techniques had been performed. These findings should lead microbiologists to focus on targeted specific real-time PCR regarding the clinical syndrome. Finally, syndrome-driven diagnosis, which consists of testing a panel of microorganisms commonly involved for each syndrome, permitted the establishment of 31 incidental diagnoses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Molecular biology has been playing an increasingly important role in the diagnosis of infectious diseases since the invention of polymerase chain reaction (PCR) in 1983 [1]. Its rapid implementation in clinical microbiology laboratories has been one of the key diagnostic tests in disciplines as diverse as virology [2], parasitology/mycology [3] and bacteriology [4]. If the traditional techniques such as axenic culture remain essential for the diagnosis of infectious diseases, molecular tools have shown an added value in the diagnosis of infections caused by fastidious bacteria [5–7], intracellular microorganisms [7, 8] and in patients receiving antibiotics [7, 9, 10]. Universal PCR targeting the sequence of ribosomal 16S rDNA was gradually established in microbiology laboratories [11], in particular to identify the causative agents of osteoarticular infections [9] or negative blood culture endocarditis [10, 12, 13]. Its contribution is undeniable today, despite the pitfalls inherent in the technique [14, 15] and a lack of sensitivity that can be criticised [16, 17]. Real-time PCR (RT-PCR) systems were subsequently developed with the advent of automation for the fluorescence detection of amplicons generated concomitantly with amplification. Molecular diagnosis had been revolutionised by the advent of this technique through two aspects: rendering speedy results (the RT-PCR process is complete in about 1 h) and reducing the risk of contamination, by amplification and detection being carried out in a closed system [4]. This makes RT-PCR the investigational method of choice in emergency diagnosis and explains its presence in structures such as point-of-care [18]. Furthermore, molecular tools have proven to be more sensitive than culture in some contexts, such as the diagnosis of osteoarticular infections caused by Staphylococcus aureus or Mycobacterium tuberculosis [16, 19]. Syndrome-driven diagnosis, which consists of testing a panel of aetiological agents frequently involved in each syndrome, has permitted a significant improvement in microbiological diagnosis [20], expanding the panel of microorganisms tested. These syndromic PCR panels, combining both RT-PCR and conventional PCR, also include emerging pathogens that have been gradually incremented, especially following molecular studies combining the amplification, cloning and sequencing of ribosomal 16S rDNA [21], to ensure the completeness of microbiological documentation. Herein, we report our experience in the use of molecular tools applied to the syndrome-driven diagnosis of infectious diseases. This 2-year retrospective study concerns samples from both patients hospitalised locally and those addressed as part of the National Reference Center activity (Coxiella burnetii, Bartonella spp., Rickettsia spp.).
Materials and methods
Samples and patients
This 2-year retrospective study includes all clinical samples addressed for bacterial DNA detection and identification from November 2011 through November 2013 and PCRs from Centre National de Réference des Rickettsies activity (detection of Bartonella spp., Coxiella burnetii, Ehrlichia, Rickettsia spp.). PCRs performed on bacterial strains (identification, virulence factor detection, typing, antibiotic resistance detection) have been excluded from the study. For each positive sample, molecular results were compared with those of an axenic culture, when available. This comparison was not performed on intracellular microorganisms due to the fastidious culture. Samples received were categorised in a syndrome when available (Online Resource 3) that generates specific RT-PCR and broad-range PCR, such as the endocarditis panel realised for each valvular resection. The panel of microorganisms tested was regularly incremented, based on epidemiological data, surveillance of microbiological laboratory results or queries from clinicians [20, 22].
DNA extraction
DNA extraction was performed on an EZ1 Advanced XL device using a Qiagen DNA tissue Kit cartridge (Qiagen, Courtaboeuf, France). From urine, bronchoalveolar lavage (BAL), vitreous punctures, sera or ethylenediaminetetraacetic acid (EDTA)-blood samples on which mycobacterial detection was not performed, 200 μL was directly eluted into 50 μL without pre-treatment. Concerning other liquid samples as for solid ones, a moiety or 200 μL of the sample was added to 200 μL of Buffer G2 and 20 μL of proteinase K (Qiagen) and heated for 2 h at 56 °C. Acid-washed glass beads (<106 μm, Sigma Aldrich, Saint-Quentin Fallavier, France) were added, and the obtained preparation was vigorously shaken using a FastPrep device (MP Biomedicals, Illkirch, France) for 20 s. After heating at 100 °C for 10 min, 200 μL was used for DNA extraction and eluted in 100-μL fractions. The quality of extraction was quantified using RT-PCR, which targeted a sequence coding for β-globin as previously described [9]. A cycle threshold (Ct) under 30 Ct was considered acceptable.
Conventional PCR and sequencing
Conventional PCR is used for targeting sequences coding for 16S rDNA, 18S rDNA (universal eukaryotes), universal fungi (CU) targeting the ITS gene and the Ureaplasma genus (Online Resource 1). A positive and a negative control (which consist of the amplification mix with 5 μL of bacterial DNA or 5 μL of H2O, respectively) were used for every ten PCRs. The amplification reaction was performed using a 2720 Thermal Cycler device as recommended by the manufacturer (Applied Biosystems, Villebon Sur Yvette, France). PCR amplicon sizes were evaluated using a QIAxcel Advanced device (Qiagen) with the QIAxcel DNA Screening Kit 2400, DNA and a molecular size marker (100 bp to 2.5 kb, Qiagen). After revelation, positive samples were sequenced using the Sanger method. Amplification products were purified by ultrafiltration using a NucleoFast filtration plate (Macherey-Nagel, Hoerdt, France) and a vacuum pump (Millipore, Molsheim, France) after the addition of 50 μL of distilled water (Gibco Life Technologies, Austin, USA). PCR products were sequenced using a BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems). The generated PCR amplicons were purified using MultiScreen plates (Merck Millipore) by exclusion chromatography on G50 Sephadex columns (Sigma-Aldrich) and then collected with a MicroAmp® Optical 96-Well Reaction plate (Applied Biosystems). Sequence analysis was performed on an ABI 3130xl Genetic Analyzer (Applied Biosystems) using DNA Sequencing Analyzing Software following the manufacturer’s recommendations (Applied Biosystems). The correction of obtained sequences was performed using the CodonCode Aligner software (http://www.codoncode.com) and then blasted against the GenBank nucleotide collection (nr/nt) or against GenBank 16S ribosomal RNA sequences (Bacteria and Archaea) (http://blast.ncbi.nlm.nih.gov). A threshold similarity value of >98.7 % was considered acceptable at the species-level identification [23].
Real-time PCR
Real-time PCRs are performed as simplex reactions. Amplification and fluorescence detection was performed on a CFX96 device following the manufacturer’s recommendations (Bio-Rad Clinical Diagnostics, Marnes-la-Coquette, France). Primers and probes used for each system are listed in Online Resource 2. The probes used in this study are based on TaqMan technology, combining a fluorochrome reporter (FAM or VIC) with a fluorochrome quencher (TAMRA or MGB). Every result for which the Ct was under 37 was considered positive. Discordant results were controlled on a second extraction if the amount of sample allowed it.
Interpretation and validation of the results
For each PCR run, whether real-time or conventional, positive and negative controls were essential for assay validation. Extracted bacterial DNA from each of the microorganisms tested was used as a positive control. In addition, negative samples were used as negative controls. As previously described, DNA extracted from tissue without infection was used as negative samples [24], tested in parallel for each PCR assay. Thus, a tissue bank was established composed of between 10 and 50 negative samples for each type of specimen and was used to validate positive results (Online Resource 4).
Results
Samples and patients
Over the 2-year study period, 32,948 clinical samples were received from 18,056 patients, on which 81,476 PCRs were performed, allowing for the establishment of 1,192 diagnoses. Samples were grouped by syndrome (Table 1). Thus, those diagnosed with Whipple’s disease (EDTA-blood, saliva, stool samples) represented 31.8 % of the samples received (10,477/32,948), followed by osteoarticular samples (4,316, 13.1 %) and those for cardiac infection diagnoses, such as EDTA-blood samples, heart valves and pericardial liquids/biopsies (3,558, 10.8 %). Following these sample types were respiratory tract samples, such as BAL or sputum (2,998, 9.1 %), cerebro-spinal fluid (CSF) (2,306, 7 %), samples for the diagnosis of sexually transmitted infections (2,009, 6.1 %), ocular samples (1,052, 3.2 %), lymph nodes (772, 2.3 %) and, finally, cerebral abscesses (130, 0.4 %). Uncategorised samples, such as abscesses, cutaneous biopsies, urine or puncture fluids, were classified as ‘others’. The samples came from the Public Assistance Hospitals of Marseille (AP-HM) (17,776, 54 %), other French hospitals (14,748, 44.8 %) and from abroad (424, 1.2 %). Concerning patients for whom several samples were received in order to diagnose the same clinical entity, only one positive sample was kept for analysis in this study.
Aetiological agents found for each syndrome
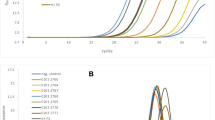
Among the 2,998 respiratory samples tested, 197 were positive (6.6 % of the total number of respiratory specimens tested). The predominant microorganisms that were identified in this series were Tropheryma whipplei (43, 21.8 % of positive samples), Mycobacterium tuberculosis (23, 11.7 %), Streptococcus pneumoniae (19, 9.6 %), Mycobacterium spp. (19, 9.6 %), Pseudomonas aeruginosa (12, 6.1 %), Mycoplasma pneumoniae (10, 5.1 %), Bordetella pertussis (9, 4.6 %) and Staphylococcus aureus (9, 4.6 %) (Fig. 1a).
Among the 3,558 samples tested for endocarditis diagnosis (EDTA-blood samples or valvular tissues), 252 were positive for 182 patients (7 % of specimens tested). The predominant microorganisms that were identified in this series were Staphylococcus aureus (24, 13.1 % of positive samples), Coxiella burnetii (23, 12.6 %), Enterococcus faecalis (20, 10.9 %), Streptococcus gallolyticus (20, 10.9 %), oral streptococci (16, 8.7 %), Tropheryma whipplei (11, 6 %), Bartonella quintana (8, 4.4 %), Streptococcus pneumoniae (8, 4.4 %) and Bartonella henselae (7, 3.8 %) (Fig. 1f).
Among the 1,052 ocular samples tested, 46 were positive for 37 patients (4.37 % of specimens tested). The predominant microorganisms that were identified in this series were Pseudomonas aeruginosa (10, 27 %), Staphylococcus epidermidis (4, 10.8 %) and Streptococcus pneumoniae (3, 8.1 %) (Fig. 1g).
Among the 4,316 osteoarticular samples tested, 370 were positive for 287 patients (8.6 % of specimens tested). The predominant microorganisms that were identified in this series were Staphylococcus aureus (122, 42.5 % of positive samples) and Kingella kingae (25, 8.7 %) (Fig. 1c).
Among the 2,306 CSF samples tested, 81 were positive for 73 patients (3.5 %). The predominant microorganisms that were identified in this series were Neisseria meningitidis (16, 21.9 % of positive samples), Streptococcus pneumoniae (12, 16.4 %), Tropheryma whipplei (9, 16.4 %), Streptococcus agalactiae (8, 11 %), Escherichia coli (4, 5.5 %), Pseudomonas aeruginosa (4, 5.5 %) and Staphylococcus aureus (3, 4.1 %) (Fig. 1e).
Among the 772 lymph nodes tested, 147 were positive for 131 patients (19 % of specimens tested). The predominant microorganisms that were identified in this series were Bartonella henselae (68, 51.9 % of positive samples), Staphylococcus aureus (14, 10.7 %), coagulase-negative staphylococci (8, 6.1 %), Mycobacterium tuberculosis (6, 4.6 %) and Streptococcus pyogenes (4, 3.1 %) (Fig. 1b).
Among the 130 cerebral abscesses tested, 61 were positive for 44 patients (47 % of specimens tested). The predominant microorganisms that were identified in this series were Staphylococcus aureus (11, 25 % of positive samples), Streptococcus intermedius (10, 22.7 %), Propionibacterium acnes (3, 6.8 %) and Mycoplasma faucium (2, 4.5 %) (Fig. 1d).
Usefulness of molecular tools
Real-time specific PCR versus conventional broad-range PCR
Among the 437 diagnoses included in this study for which both conventional and targeted RT-PCR had been performed, 275 were established by RT-PCR only (62.9 %). A total of 28 % of these diagnoses included endocarditis. In this clinical context, microorganisms for which only RT-PCR was positive with a negative conventional amplification were Streptococcus gallolyticus (16/20, 80 %), Staphylococcus aureus (16/24, 67 %), Enterococcus faecalis (11/20, 55 %), Coxiella burnetii (16/23, 70 %), Bartonella henselae (4/7, 57.1 %), Bartonella quintana (3/8, 37.5 %), Enterococcus faecium (2/2, 100 %) and Streptococcus pneumoniae (3/7, 42.8 %). RT-PCR also showed a better detection rate for the diagnosis of osteoarticular infections, for which the sensitivity fell below 50 % (30/175), in particular in those due to Staphylococcus aureus, for which only 19 (16.2 %) were positive using conventional broad-range PCR as well as only seven of the 26 infections due to Kingella kingae (27 %). The majority of meningitis diagnoses (51/73, 70 %) were established using specific RT-PCR. Of the 12 cases for which conventional broad-range PCR had been performed, only four were positive (33 %). Finally, of the 72 lymph nodes that were positive for Bartonella henselae using specific RT-PCR, only 50 % were also positive using broad-range PCR; the result was similar for Francisella tularensis (four diagnoses, 25 % positive using 16S rDNA amplification).
Broad-range conventional PCR remains useful when specific RT-PCR for particular targets are not available in our microbiology lab, as was the case with 109 of the 287 osteoarticular infections (38 %), 56 of the 197 respiratory samples (28 %), 24 of the 131 adenitis samples (18.3 %), 41 of the 183 endocarditis samples (22.4 %), 19 of the 73 meningitis samples (26 %) and 19 of the 44 cerebral abscesses (43.2 %). Moreover, complementarity between these two techniques allowed for the identification of several aetiological agents for 16 polymicrobial samples, which mainly concern BAL and cerebral abscesses (five for both) (Table 2). Finally, RT-PCR failed to detect three endocarditis (two due to Enterococcus faecalis and one due to Streptococcus mitis) and one osteoarticular infection due to Staphylococcus aureus that were all positive by broad-range conventional PCR.
Benefits of molecular tools compared to axenic culture
Among the 1,192 positive samples for which activity is independent of the National Reference Center, 25 % were sterile in culture and 6 % yielded a different microorganism than that found by PCR (data not shown). Culture-negative infections due to cultivable bacteria represented 60.5 % of meningitis, 37.7 % of osteoarticular infections and 59.5 % of endocarditis diagnoses (Table 1). Culture-negative osteoarticular infections were mostly Staphylococcus aureus (37/88, 42 %) and Kingella kingae (8/88, 9.1 %). For blood culture-negative endocarditis, excluding strict intracellular bacteria, Streptococcus gallolyticus predominated (12/48, 22.4 %), followed by Staphylococcus aureus (10/48, 20.4 %), Enterococcus faecalis (7/48, 14.2 %) and Streptococcus pneumoniae (4/48, 8.1 %). Six tuberculous lymphadenopathies were detected by specific RT-PCR, which permitted early diagnosis by culture, of which one node remained sterile. Moreover, nine anaerobic agents were detected using conventional broad-range PCR on osteoarticular samples, of which four did not grow. Finally, four fastidious or anaerobic agents were detected in cerebral abscesses, of which only three were cultivated (Table 3).
Contribution of a systematic syndromic PCR panel and incidental diagnosis
Incidental diagnosis concerned samples collected outside of the AP-HM, for which a systematic panel of PCR regarding the clinical syndrome was performed, regardless of the initial request. Thus, the use of a systematic syndromic PCR panel permitted the identification of an aetiological agent for five cerebral abscesses, 15 osteoarticular infections, eight endocarditis, one sexually transmitted disease (STI), one tuberculosis lymphadenitis and one pericarditis (Table 4).
Usefulness of conventional fungal and eukaryotic amplification
Fungal amplification targeting the 28S ribosomal subunit gene allowed for the diagnosis of 14 infections, four of which were not detected by culture methods . These diagnoses include six from respiratory samples, three osteoarticular infections and two ophthalmic infections. The amplification of the eukaryotic 18S ribosomal subunit gene allowed for the diagnosis of an ocular infection due to Acanthamoeba castellanii.
Discussion
Herein, we report our experience regarding the diagnosis of infectious diseases using molecular tools. We are confident in our results; using PCR routinely for the last decade has allowed us to assess many PCR systems, which enabled us to confirm even our most unexpected results (Online Resources 1 and 2). The shift to the syndrome-driven diagnosis, which consists of testing a panel of microorganisms frequently involved in a clinical context, ensures the completeness of the microbiological documentation.
Molecular tools have shown their benefits concerning the diagnosis of infections caused by fastidious microorganisms. Thus, 27 % of the endocarditis cases included in this series were due to strict intracellular bacteria, as were 57 % of the adenitis diagnoses. Likewise, osteoarticular infections caused by Kingella kingae, a fastidious bacterium requiring culture enrichment and prolonged incubation [25], is more likely to be diagnosed by PCR, as shown by the five isolates cultured among the 26 diagnoses established. Finally, among other fastidious bacteria (bacteria of the HACCEK group, anaerobes and deficient streptococci) identified in this work, 18 had not been cultured (Table 3). The complementarity between molecular and culture-dependent techniques is illustrated by the proportion of positive samples for which a culture is negative, up to 25 %. Bacterial dormancy is a possible explanation for the negative cultures, especially in cases of osteitis with low inoculum. Furthermore, antibiotic therapy administered before sampling, particularly concerning endocarditis before surgery, is common. RT-PCR also permitted the early diagnosis of 64 mycobacterial infections (45 pulmonary forms, 11 osteoarticular forms and eight lymphadenitis) before culture, which may change, as spectacular improvements have recently been reported [26].
The difference in sensitivity between specific RT-PCR and broad-range PCR in this study is dramatic. Indeed, the use of conventional PCR would have only allowed us to diagnose 53.6 % of endocarditis included in this study, along with 51.2 % of the osteoarticular infections, with detection rates estimated at 38.3 % and 20 %, respectively.. However, conventional broad-range PCR allowed us to identify bacteria for which specific RT-PCR systems were not available, which includes 22.3 % (266/1,192) of infections diagnosed in this work, particularly osteoarticular infections, for which a large variability of aetiological agents can be involved. Its contribution is essential to identify emerging, rare or unexpected pathogens, as illustrated by the 125 different microorganisms found in this work (Fig. 2). Even if polymicrobial infections were one of the pitfalls of conventional broad-range PCR [24], its association with pathogen-specific RT-PCR permitted the diagnosis of 16 polybacterial infections (Table 2).
The syndrome-driven diagnosis, which groups a panel of microorganisms to test, allowed us to diagnose 31 infections, including 15 osteoarticular, eight endocarditis and five cerebral abscesses. These benefited from the incrementing of specific PCRs targeting Mycoplasma faucium and Streptococcus intermedius following a multiple 16S rDNA sequencing study [21, 27] (Table 2, Online Resource 3). Furthermore, the high prevalence of Tropheryma whipplei in BAL (21.8 %), which was initially tested prospectively, should lead us to add this targeted RT-PCR to the respiratory tract infection syndromic PCR panel.
In conclusion, this study focuses on the complementarity of molecular tools with culture techniques, in particular for fastidious or intracellular microorganisms, and antibiotic administration before sampling. This study also adds value to the diagnosis of meningitis, endocarditis, osteoarticular infections and adenitis [28]. The dramatic lack of sensitivity of conventional broad-range PCR reported in this work should lead microbiologists to perform in priority targeted specific RT-PCR regarding the clinical syndrome.
References
Mullis K, Faloona FA, Scharf SJ, Saiki RK, Horn GT, Erlich H (1992) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Biotechnol Ser 17
Mackay IM, Arden KE, Nitsche A (2002) Real-time PCR in virology. Nucleic Acids Res 30:1292–1305
Bell AS, Ranford-Cartwright LC (2002) Real-time quantitative PCR in parasitology. Trends Parasitol 18:337–342
Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JDC, Wengenack NL, Rosenblatt JE, Cockerill FR 3rd, Smith TF (2006) Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19:165–256
Brouqui P, Raoult D (2001) Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 14:177–207
Fenollar F, Raoult D (2004) Molecular genetic methods for the diagnosis of fastidious microorganisms. APMIS 112:785–807
Fenollar F, Raoult D (2007) Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents 30:S7–S15
Houpikian P, Raoult D (2005) Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 84:162–173
Fenollar F, Lévy PY, Raoult D (2008) Usefulness of broad-range PCR for the diagnosis of osteoarticular infections. Curr Opin Rheumatol 20:463–470
Rovery C, Greub G, Lepidi H, Casalta JP, Habib G, Collart F, Raoult D (2005) PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J Clin Microbiol 43:163–167
Sontakke S, Cadenas MB, Maggi RG, Diniz PPV, Breitschwerdt EB (2009) Use of broad range 16S rDNA PCR in clinical microbiology. J Microbiol Methods 76:217–225
Casalta JP, Gouriet F, Thuny F, Bothelo E, Lepidi H, Fournier PE, Habib G, Guidon C, Collard F, Raoult D (2009) Standardisation et prise en charge multidisciplinaire des endocardites. Stratégie du CHU de Marseille. Antibiotiques 11:81–89
Goldenberger D, Künzli A, Vogt P, Zbinden R, Altwegg M (1997) Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol 35:2733–2739
Janda JM, Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764
Millar BC, Xu J, Moore JE (2002) Risk assessment models and contamination management: implications for broad-range ribosomal DNA PCR as a diagnostic tool in medical bacteriology. J Clin Microbiol 40:1575–1580
Fenollar F, Roux V, Stein A, Drancourt M, Raoult D (2006) Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol 44:1018–1028
Fihman V, Hannouche D, Bousson V, Bardin T, Lioté F, Raskine L, Riahi J, Sanson-Le Pors MJ, Berçot B (2007) Improved diagnosis specificity in bone and joint infections using molecular techniques. J Infect 55:510–517
Cohen-Bacrie S, Ninove L, Nougairède A, Charrel R, Richet H, Minodier P, Badiaga S, Noël G, La Scola B, de Lamballerie X, Drancourt M, Raoult D (2011) Revolutionizing clinical microbiology laboratory organization in hospitals with in situ point-of-care. PLoS One 6:e22403
Lecouvet F, Irenge L, Vandercam B, Nzeusseu A, Hamels S, Gala JL (2004) The etiologic diagnosis of infectious discitis is improved by amplification-based DNA analysis. Arthritis Rheum 50:2985–2994
Gouriet F, Raoult D (2009) Diagnostic microbiologique: du diagnostic par étiologie au diagnostic par syndrome. Antibiotiques 11:37–48
Al Masalma M, Armougom F, Scheld WM, Dufour H, Roche PH, Drancourt M, Raoult D (2009) The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin Infect Dis 48:1169–1178
Parola P, Colson P, Dubourg G, Million M, Charrel R, Minodier P, Raoult D (2011) Letter to the editor. Group A streptococcal infections during the seasonal influenza outbreak 2010/11 in South East England. Euro Surveill 16(11). pii: 19816
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152
Lévy PY, Fenollar F (2012) The role of molecular diagnostics in implant-associated bone and joint infection. Clin Microbiol Infect 18:1168–1175
Yagupsky P, Dagan R, Howard CW, Einhorn M, Kassis I, Simu A (1992) High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J Clin Microbiol 30:1278–1281
Ghodbane R, Raoult D, Drancourt M (2014) Dramatic reduction of culture time of Mycobacterium tuberculosis. Sci Rep 4:4236
Mishra AK, Fournier PE (2013) The role of Streptococcus intermedius in brain abscess. Eur J Clin Microbiol Infect Dis 32:477–483
Angelakis E, Roux V, Raoult D, Rolain JM (2009) Real-time PCR strategy and detection of bacterial agents of lymphadenitis. Eur J Clin Microbiol Infect Dis 28:1363–1368
Funding
This work was funded by IHU Méditerranée Infection.
Conflict of interest
None of the authors have a conflict of interest.
Ethical approval
Permission from the local ethics committee of the IFR48 (Marseille, France) was obtained under agreement 09-022.
Author information
Authors and Affiliations
Corresponding author
Additional information
Anne-Sophie Morel and Grégory Dubourg contributed equally to this work.
Electronic supplementary material
Below are the links to the electronic supplementary material.
Online Resource 1
(PDF 326 kb)
Online Resource 2
(PDF 363 kb)
Online Resource 3
(PDF 16 kb)
Online Resource 4
(PDF 92 kb)
Rights and permissions
About this article
Cite this article
Morel, AS., Dubourg, G., Prudent, E. et al. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis 34, 561–570 (2015). https://doi.org/10.1007/s10096-014-2263-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2263-z