Abstract
The effects of the secondary metabolite biosynthesis on the metabolism and morphology of the Monascus purpureus were investigated in this study. Hypha and septum length became longer after deletion of genes pigR and pksCT in M. purpureus LQ-6 by Agrobacterium tumefaciens-mediated transformation technology, highly branched hyphae, much smaller and freely dispersed mycelial pellets were observed in M. purpureus. Compared with that in the wild-type, the level of intracellular NADH and NADPH was almost constant in M. purpureus ΔpigR at 4 days, but the NADH and NADPH levels decreased by 1.58-fold and 3.71-fold in M. purpureus ΔpksCT. The present study can not only provide a kind of strategy to improve the Monascus pigments production, but also provide theoretical support for the further study of relationship between the secondary metabolites, metabolism and morphological change.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fungus Monascus spp. has been well-known in many oriental countries for centuries, especially in China, Japan, and Korea. Monascus spp. is conventionally cultured on steamed white polished rice to produce a fermented meal known as red mold rice (RMR) in the USA, Hongqu in China, and Koji and Red Koji in Japan. There are hundreds of scientific studies that indicate that RMR contains many kinds of beneficial compounds, such as natural pigments (used as food colorants), monacolin K or lovastain (HMG-CoA reductase inhibitor), and gamma-amino butyric acid (GABA, a kind of hypotensive agent) (Patakova, 2013; Shao et al., 2014). However, the usage of RMR also incurs controversy since the discovery of citrinin (a kind of mycotoxin, shown to be nephrotoxic in animals) production by Monascus strains. Although there are no strict rules limiting the upper amount of citrinin in RMR in China, a limit of 2000 ppb (2000 µg/Kg) in RMR was established in Europe according to UC Commission regulation No. 212/2014, 50 ppb in South Korea (Kim et al., 2007), and 200 ppb in Japan (Fu et al., 2007). However, according to Chinese National Standard GB 1886.181-2016, the upper limit of citrinin was determined to be 0.04 ppm/U in MPs.
Pigments, monacolin K, and citrinin are the three most famous polyketide (PK) secondary metabolites produced by filamentous fungi Monascus strains, which were catalyzed by polyketide synthase, composed of catalytic domains consisting of ketosynthase (KS), acyltransferase (AT), product template (PT), acyl carrier protein (ACP), and C-methyltranferase (MT) (Balakrishnan et al., 2013). MPs are a mixture of azaphilones mainly composed of yellow pigments (ankaflavin and monascin), orange pigments (monascorubrin and rubropunctatin), and red pigments (monascorubramine and rubropunctamine) (Kim and Ku 2018; Wang et al., 2015), and the related metabolic pathway showed that yellow pigments are formed from orange pigments by reduction, and the orange colors could convert into red when oxygen atoms are replaced by an amino group.
One of the main challenges in the production of fungal metabolites by filamentous fungi such as Monascus is the control of mycelial morphology, which attached great importance to the production of metabolites (Lv et al., 2017). Hyphae of filamentous fungi in submerged batch-fermentation (SBF) mainly exist in three different morphology, including free mycelia, mycelial pellets mycelial clumps (Zhang et al., 2015). Filamentous fungi morphology are much affected by fermentation conditions and genetic factors, such as pH, shaking speed (Ibrahim, 2015; Lv et al., 2017), oxygen diffusion (Vecht-Lifshitz et al., 1990), exogenous additive such as cyclic AMP (Lai et al., 2011) and nonionic surfactant Triton X-100 (Chen et al., 2018). The nonionic surfactants were correlated with pigment production and morphology in SBF, which resulted in a higher level of MPs production and changed the mycelia morphology (Yang et al., 2019). In the production of Monascus red pigment, the maximum production of MPs was obtained with high agitation rates, and shorter branches were produced in Monascus mycelium (Kim et al., 2002). To note, many scholars have studied the relationship between mycelial morphology and pigment, but whether the effect of citrinin biosynthesis on mycelial morphology is the same as that of pigment has not been investigated. Furthermore, the effects of pigment synthesis and citrinin metabolism on Monascus metabolism such as cofactor metabolism are still unclear.
It is well-known that three secondary metabolites—pigments, monacolin K, and citrinin—share a biosynthetic pathway before a certain branch point, synthesized by acetyl CoA and malonyl-CoA, especially for pigment and citrinin (Fig. 1). Although many researchers have found that disruption of citrinin or monacolin K biosynthesis could increase the production of pigment, the relationship between the secondary metabolite biosynthesis and other intracellular metabolism (such as the glycolysis pathway and cofactor generation) and the effect of the secondary metabolites on the cell growth and morphology and physiology of hypha are still unclear. Hence, the aim of this work was to investigate the effect of the secondary metabolite biosynthesis on the metabolism and morpholog of the M. purpureus strain by disruption of the biosynthetic pathway of pigments and citrinin using Agrobacterium tumefaciens-mediated transformation (ATMT) technology.
Materials and methods
Microorganisms, cultivation, and materials
Monascus purpureus LQ-6 (CCTCC M 2018600, China Central for Type Culture Collection (CCTCC), Wuhan, China), as the parent strain, was isolated from red mold rice obtained from a market in China. M. purpureus strains were cultured on potato dextrose agar (PDA, potato (200 g), dextrose (20 g), and agar (20 g) in 1000 mL pure water) medium with the addition of 50 μg/mL hygromycin or G418 as required at 30 °C. Escherichia coli DH5α cells were grown in Luria–Bertani (LB, tryptone (10 g/L), yeast extract (5 g/L), NaCl (10 g/L)) medium or solid medium (addition of 20 g/L agar) with the addition of 50 μg/mL kanamycin as required at 37 °C. The strains and plasmids used in this study are listed in Table S1.
Construction of pigR and pksCT deletion vector
As shown in Fig. S1, homologous recombination was applied for knocking out the pigR gene in M. purpureus LQ-6. The primers used in this study are listed in Table S2. There are two steps to construct the pigR disruption vector, as follows.
To construct the pigR-deleted cassette, a 1.54 kb 5′ homologous arm and a 1.53 kb 3′ homologous arm (named 5′-UTR and 3′-UTR, respectively) were amplified from M. purpureus LQ-6 genomic DNA by the polymerase chain reaction (PCR) using the primers pigR-up-R, pigR-up-F, and pigR-dn-R, pigR-up-F, respectively. A 2.1 kb fragment containing the G418 gene from the plasmid pXS (the primers G418-R, G418-F) was fused with 5′-UTR and 3′-UTR by overlap PCR to generate the pigR-deleted cassette (Ning et al., 2017).
The pigR-deleted cassette obtained from the first step was cloned into the corresponding sites of the pCAMBIA3300 binary vector linearized by double digestion with EcoR I and Hind III using a one-step cloning kit (Vazyme Biotech Inc., Nanjing, China). Finally, the pigR deletion vector named pCAMBIA3300-ΔpigR was formed(Xie et al., 2013).
To construct the pksCT disruption vector, a similar strategy as above was used. In addition, the selection marker for pksCT was hph, which was amplified from the plasmid pAN7-1with a primer pair hph-F and hph-R.
Transformation of M. purpureus
ATMT technology was used to transform the disruption vector, according to Shao et al.(Shao et al., 2009). M. purpureus LQ-6 was cultured on a Potato Dextrose Agar (PDA) plate for 10 days at 30 °C, and then 5 mL of sterile water was added to the PDA plate with a fully-grown culture of M. purpureus LQ-6. The suspension was filtered through two layers of sterile lens paper to collect the spores. Then, A. tumefaciens EHA105 containing recombinant plasmid pCAMBIA3300-ΔpigR was inoculated for 2 days at 28 °C in minimal medium (2 g/L glucose, 2.05 g/L K2HPO4, 1.45 g/L KH2PO4, 0.01 g/L CaCl2, 0.6 g/L MgSO4, 0.3 g/L NaCl, 0.5 g/L (NH4)2SO4, pH = 7.0, supplemented with 10 mL 0.01% (v/v) FeSO4) supplemented with kanamycin (50 μg/mL). After dilution to an optical density (OD) of 0.15 at 600 nm with induction medium (2 g/L glucose, 1.84 g/L K2HPO4, 1.45 g/L KH2PO4, 0.01 g/L CaCl2, 0.6 g/L MgSO4, 0.3 g/L NaCl, 0.5 g/L (NH4)2SO4, 5 g/L glycerol, pH = 4.9, supplemented with 10 mL 0.01% FeSO4, 10 mL 100 g/L MES (pH 5.5), and 5 mL 200 mM acetosyringone), the cell suspension of A. tumefaciens EHA105 needed an additional 6 h growth under the same conditions. Next, the A. tumefaciens cell suspension was mixed with spore pellets of M. purpureus LQ-6 at a concentration of 106 spores/mL, and the mixture (200 μL) was plated on the coculture-inducing medium (1 g/L glucose, 1.84 g/L K2HPO4, 1.45 g/L KH2PO4, 0.01 g/L CaCl2, 0.6 g/L MgSO4, 0.3 g/L NaCl, 0.5 g/L (NH4)2SO4, 5 g/L glycerol, 20 g/L agar, supplemented with 10 mL 0.01% (v/v) FeSO4, 10 mL 100 g/L MES (pH 5.5), and 10 mL 200 mM acetosyringone) covered with cellophane. After co-cultivation for 4 days at 25 °C, the cellophane was removed, and an empty sterile culture dish with poured PDA containing 50 μg/mL G418 for selecting transformants and 200 μg/mL cephalosporin for killing the A. tumefaciens cells was used. Following cultivation for 2 days at 25 °C, single colonies were picked up and cultured on PDA supplemented with G418. If these colonies still grew on the selective medium, they were regarded as transformants.
For deletion of gene pksCT (50 μg/mL hygromycin as the selection agent) and double deletion of genes pigR and pksCT (disruption of gene pksCT in the background of recombinant stain M. purpureus ΔpigR), the same method described above was performed.
Confirmation of recombination strains
After Agrobacterium tumefaciens-mediated transformation of the disruption vectors, PCR analysis was subjected to identify transformants. For PCR analysis, genomic DNA was isolated from transformant mycelia using the CTAB method(Shao et al., 2009). Primers pigR-T-F/R, pigR-half-F, and G418-T-R were used to confirm the deletion fragment of pigR and the double deletion fragment of pigR and pksCT. The deletion fragment in the genomic DNA of the hygromycin-resistant transformants was amplified using primers CT-T-F/R, CT-half-F, and hyh-T-R. The recombinant strains were named M. purpureus ΔpigR (pigR-deleted strain), M. purpureus ΔpksCT (pksCT-deleted strain), and M. purpureus ΔpksCT:pigR (pigR and pksCT-double deleted strain), respectively.
Submerged batch-fermentation
Sterile water (10 mL) was added to the PDA medium with a fully-grown culture of M. purpureus and aseptically scraped using a sterile inoculating loop to obtain a spore suspension. For SBF, 5 mL of the spore suspension (106 spores/mL) was inoculated into 45 mL liquid-state fermentation medium in 250 mL conical flasks at 30 °C while shaking at 150 rpm in the dark for 252 h (Liu et al., 2019; Terán et al., 2018). Fermentation kinetics assays were conducted in 1000 mL conical flasks containing 200 mL SBF medium.
Image analysis
For the physiological characteristics assay, a 5 µL spore suspension was inoculated into the middle of a PDA plate (diameter = 90 mm) at 30 °C to analyze the colony size and spore development rate. For 5 days, SBF samples were taken to observe the morphology of mycelial pellets. All of them were monitored by a digital camera (Canon EOS 80D, Japan).
For the microscopy assay, the hyphae of strains were taken using a sterile inoculating loop from the cultured PDA plate for 3 and 5 days, respectively. Afterwards, the hyphae were re-suspended in sterile water and imaged using a Motic compound microscope (BA200, Germany).
Metabolites analytic methods and calculation
The fermentation broth was centrifuged at 8000 rpm for 10 min to collect mycelia and supernatant, respectively. For total biomass determination, the collected mycelia were washed three times with distilled water and dried at 60 °C. The total biomass concentration was expressed as dry cell weight (DCW) per unit volume of fermentation medium. The supernatant was diluted to measure the residual glucose concentration using the standard 3,5-dinitrosalicylic acid (DNS) method. After SBF finished, the collected mycelia were soaked in 5 volumes of 70% (v/v) ethanol at 60 °C for 2 h for determination of the intracellular pigment production. The fermentation supernatant was directly measured for analysis of extracellular pigments. The concentration of MPs was analyzed by measuring the corresponding absorbance by a UV–visible spectrophotometer (UV-752 N), at 505 nm for red pigments, 470 nm for orange pigments and 420 nm for yellow pigments, respectively. The total MPs concentrations include intracellular pigment production and exMPs production (Chen et al., 2017).
For analysis of monacolin K and citrinin production, 5 mL of the fermentation medium was inoculated into 20 mL of 75% (v/v) methanol at 60 °C for 2 h, followed by cooling to room temperature and allowing to settle overnight. After centrifuging at 10,000 rpm for 5 min and filtering through a 0.22 µm filter, the supernatant was assessed by high performance liquid chromatography (HPLC, LC-30A, Shimadzu) on a reverse phase C18 column (5 µm, 150 × 4.6 mm) (Zhang et al., 2017). Monacolin K production was determined with methanol/water (70:30, pH adjusted to 3 with orthophosphoric acid) as the mobile phase, 1.0 mL/min flow rate at 25 °C, and 10 µL injection volume, and the eluate was monitored by a UV detector at a wavelength of 237 nm(Lin et al., 2018). To determine the concentration of citrinin, a mixture of acetonitrile, isopropanol, and 0.08 mol/L orthophosphoric acid at a ratio of 35:10:55 (v/v) as the mobile phase, with a 1.0 mL/min flow rate at 25 °C and injection volume of 10 µL was examined using a fluorescence detector (Prominence RF-20A/20Axs) at a 331 nm excitation wavelength and 500 nm emission wavelength (according to the Chinese National Standard GB 5009.222-2016).
Intracellular concentrations of NADH and NADPH were detected via Amplite™ Colorimetric NAD/NADH and NADP/NADPH Assay Kits (Beijing Solarbio Science & Technology Co., Ltd., China). Mycelium were taken from the batch-fermentation and then rapidly ground into a powder using liquid nitrogen. The mycelium powder (0.1 g, wet weight) was transferred quickly into a sample tube for measurement(Huang et al., 2017).
Each experiment was repeated at least three times. Numerical data are presented as the mean ± standard deviation (SD).
Accession numbers
The genome sequences have been deposited into the NCBI database with an accession number of PRJNA503091.
Results and discussion
Sequence determination and bioinformatics analysis
There are many kinds of Monascus, including M. ruber, M. purpureus, M. fuliginosus, M. aurantiacus, M. pilosus and so on. To determine the species, strain LQ-6 isolated naturally was identified by ITS4/5. After purification of the PCR product, the ITS fragment of strain LQ-6 was sequenced and was homologous to multiple strains of Monascus, especially M. purpureus. Furthermore, the phylogenetic tree by selecting strains with high homology and other strains of the same genus was constructed (Fig. 2A). The phylogenetic tree indicated that the strain LQ-6 (named unk in Fig. 2A) displayed high homologies with M. purpureus, M. ruber, M. pilosus, and M. fuliginosus, especially M. purpureus Z1 (97% query cover and 99.77% identity from NCBI-BLASTN). The strain LQ-6 was named as M. purpureus LQ-6.
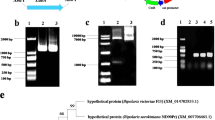
Gene pigR, is a pathway-specific regulatory gene for MPs biosynthesis, which is of great significance in MPs biosynthesis pathway(Xie et al., 2013). Gene pksCT is highly correlated with citrinin production, which is polyketide synthase gene responsible for citrinin biosynthesis (Shimizu et al., 2005). In this study, whole genome sequencing was performed to unearth gene pigR and pksCT in M. purpureus LQ-6, and the biosynthetic pathway of pigment and citrinin was blocked by disrupting the gene pigR and pksCT, respectively. Genes monacsus_3525 and monacsus_3836 were unearthed after sequencing the whole genome of M. purpureus LQ-6 and showed significant similarity to pigR and pksCT, respectively. The nucleotide sequence of gene monacsus_3525 was 3634 bp in length and encoded by 573 amino acids. A database search with NCBI-BLASTP suggested that it has 92.48% similarity with the pigment biosynthesis activator pigR (encoded by 554 amino acids) of the pigment biosynthesis cluster from Monascus ruber M7 (protein ID = AGL44390.1). Based on the results of NCBI-BLASTP, the phylogenetic tree of gene monacsus_3525 was constructed. It demonstrated that the gene monacsus_3525 displayed high homologies with pigR in M. ruber (query cover = 100%, identity = 92.48%, bootstrap value = 78), pigment biosynthesis gene cluster in M. pilosus, and then citrinin biosynthetic gene cluster in M. ruber (identity = 46.10%) (Fig. 2B). In addition, there was almost no pigment produced after deletion of gene Monascus_3525 (Figs. 3 and 5C). Gene pksCT was highly conserved in different citrinin-producing Monascus strains (Chen et al., 2008; Fu et al., 2007), encoding polyketide synthase, which is responsible for citrinin biosynthesis (Jia et al., 2010). The nucleotide acid sequence of gene monacsus_3836 was used as a query to perform BLASTN in the NCBI database. The results revealed that gene monacsus_3836 had 100% similarity to the gene pksCT from M. purpureus (sequence ID: AB167465.1) (Fig. S2). Furthermore, deletion of the gene Monascus_3836 in M. purpureus LQ-6 resulted in no citrinin production (Fig. S4). The above results and analysis indicate that the genes Monascus_3525 (pigR) and monscsus_3836 (pksCT) play a vital role in pigment and citrinin biosynthesis of M. purpureus LQ-6, respectively.
Colonial morphologies of wild-type strain M. purpureus LQ-6 and recombination strains cultivated on PDA plates at 30 °C for 5 days (A). Relative colony diameter of different strains cultivated on PDA plates on the 5th, 6th, and 7th days, with the colony diameter of M. purpureus LQ-6 as a reference (B). Mycelial morphologies of M. purpureus LQ-6 and recombination strains fermented at 150 rpm for 5 days in SBF medium (C)
However, there was no monacolin K (acid form and lactone form) produced by solid-state fermentation using M. purpureus LQ-6 (Fig. S5). It has been reported that some M. purpureus strains lack the monacolin K synthesis locus and are incapable of producing monacolin K (Kwon et al., 2016). Then, it’s speculated that the monacolin K biosynthetic gene cluster in M. purpureus LQ-6 was incomplete or lacking. The results of whole genome sequencing of M. purpureus LQ-6 showed that just genes Monascus_1765, Monascus_1766, Monascus_1767, Monascus_1768, Monascus_1769, Monascus_1770, and Monascus_1771 were located in the monacolin K biosynthetic gene cluster, and the protein homology was low, except for Monascus_1766 with 83.78% identity to the protein mkC and Monascus_1765 with 74.49% identity to the protein mkB from Monascus pilosus by BLASTP analysis. Through sequence analysis and solid-state fermentation, it’s concluded that there was no monacolin K produced by M. purpureus LQ-6, resulting from the biosynthetic gene cluster being incomplete. In addition, different strains of Monascus should be selected to study different secondary metabolites. Generally, Monascus fuliginosus, Monascus ruber and Monascus pilosus are considered to have strong production capacity of monacolin K, which were often used in monacolin K related research. M. purpureus has strong ability of producing pigment, but also with citrinin. Furthermore, given that many researches focus on the aspect of food safety to decrease the citrinin production by M. purpureus fermentation, then this strain LQ-6 was finally engineered. Besides, this study mainly focus on the morphology and metabolism in submerged fermentation after disrupting pigment and citrinin biosynthesis pathways.
Effect of biosynthesis of pigment and citrinin on morphology of M. purpureus LQ-6
In SBF, the production of secondary metabolites is strongly associated with the fungal morphology and growth (Chen et al., 2018; Sun et al., 2018). We speculated that disruption of the biosynthetic pathways of pigments and citrinin would change the development and cellar morphology in M. purpureus LQ-6. In this study, the changes in spore development, hyphal length and branching, and mycelial pellet morphology were analyzed. The colony phenotypes of LQ-6 and recombination strains incubated on PDA at 30 °C for 5 days are shown (Fig. 3A), and the order of spore germination rate was M. purpureus ΔpigR, M. purpureus ΔpksCT:pigR, M. purpureus LQ-6, and M. purpureus ΔpksCT. In addition, the hyphae of M. purpureus ΔpigR and M. purpureus ΔpksCT:pigR were white, which resulted from blocking the pigment biosynthesis pathway after deletion of gene pigR. Lv et al. have been reported the hyphal diameter was highly correlated with the biosynthesis of the MPs (Lv et al., 2017). The colony diameter of LQ-6, ΔpksCT, ΔpigR and ΔpksCT:pigR was 31.75, 27.50, 33.25and 32.75 mm, respectively. During the process of cultivation, the diameter of M. purpureus ΔpksCT was smaller than that of the wild-type strain, but the diameters of M. purpureus ΔpigR and M. purpureus ΔpksCT:pigR were bigger than that of the wild-type strain, and the differences in colony diameter were more obvious with the development of culture process (Fig. 3B). These phenomena illustrated that the genes pigR and pksCT have a great impact on spore germination and development, and blocking pigment biosynthesis can increase the cell growth rate, whereas it is reduced after disrupting citrinin biosynthesis. After the pigR gene was knocked out, the growth of Monascus did not inhibited, and the growth rate was higher than that of the parental strain LQ-6, indicating that the pigR gene was not involved in the primary metabolism synthesis, only related to MPs biosynthesis. Based on aboved results, observing the mycelial morphology and the measuring hyphal diameter can preliminary predict the production of Monascus secondary metabolites.
In SBF, two main morphologies were observed, including pellets and free mycelium. In addition, the morphology of filamentous fungi in SBF plays a significant role in metabolites yield as well as productivity (Ibrahim, 2015). In this study, mycelial pellets were formed by development from the spores and hyphae of the wild-type strain LQ-6 at 150 rpm for 5 days in SBF medium, as well as M. purpureus ΔpksCT, except the surface color of the pellets was redder (Fig. 3C). However, compared to the mycelial pellets of the wild-type strain, they were much smaller and more freely dispersed in the liquid fermentation medium after deletion of gene pigR in M. purpureus LQ-6. Furthermore, the number of cleistothecium (seen at 3 days) and hypha length (seen at 5 days) of M. purpureus were both increased by deletion of genes pigR and pksCT, especially in M. purpureus ΔpksCT (Fig. 4). Additionally, the septum of hypha became longer in M. purpureus ΔpksCT and M. purpureus ΔpigR than that in the wild-type strain. It reported that the extent of increase of hyphae branches was used as an indicator of the cell growth rate of the fungi (Papagianni, 2004). In this study, highly branched hyphae were observed in M. purpureus ΔpigR but did not appear in M. purpureus ΔpksCT, which was consistent with the above-mentioned results. There may be competitive utilization of some nutrients in the biosynthesis of MPs and the growth of Monascus, which leads to the inhibition of pigments biosynthesis on the growth of Monascus. These findings revealed that not only changing the morphology of Monascus can affect the pigments yield, but also blocking the synthesis of pigment and citrinin can affect the morphology and growth rate of M. purpureus.
Effect of biosynthesis of pigment and citrinin on the metabolism of M. purpureus LQ-6
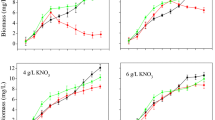
To evaluate the effect of pigments and citrinin biosynthesis on the metabolism of M. purpureus, the corresponding biosynthetic pathways were disrupted by deletion of genes pigR and pksCT. From the analysis of fermentation kinetics of various strains (Fig. 5), the recombination strains (M. purpureus ΔpigR, M. purpureus ΔpksCT, and M. purpureus ΔpksCT:pigR) showed a higher glucose consumption rate compared to the wild-type strain M. purpureus LQ-6 at the fermentation prophase, especially M. purpureus ΔpigR. In addition, the wild-type strain showed a long lag phase (glucose consumption rate was 0.10 g/L h for 36 h), which did not appear in the fermentation kinetics of the three recombination strains with a similar glucose consumption rate (Fig. 5A). These results revealed that deletion of genes pigR and pksCT can reduce the fermentation lag phase in M. purpureus LQ-6. In addition, investigation of the cell growth coupled with the glucose consumption in the fermentation process (Fig. 5B) showed that the cell growth rate slightly increased after deletion of gene pigR and double knockout of genes pigR and pksCT, which resulted in 1.04-fold and 1.02-fold times the DCW of the wild-type, respectively. However, the pksCT-deleted strain decreased to 1.04-fold times the DCW of the wild-type and a lower cell growth rate. It’s speculated that the synthesis of MPs can affect the growth and glucose consumption of Monascus spp., which is consistent with a previous study (Xie et al., 2015).
Submerged batch-fermentation kinetics of M. purpureus LQ-6 and recombination strains. Glucose consumption (A), cell growth (B), pigment production (C), and citrinin production (D). Submerged batch-fermentations were studied in 1000 mL conical flasks containing 200 mL SBF medium at 150 rpm in the dark for 252 h
The secondary metabolites biosynthesis pathway is described in Fig. 1. The monacolin K biosynthesis pathway was incomplete because of the lack of one or more key genes in the gene clusters in M. purpureus LQ-6; gene pigR-regulated pigments biosynthesis and gene pksCT catalyze citrinin production. Xie et al. reported the production of citrinin was dramatically improved by 1.60-fold after deletion of the gene pigR (Xie et al., 2013). In addition, blocking of the production of citrinin resulted in 40% more pigment in M. aurantiacus Li AS3.4384 (Ning et al., 2017). Similarly, deletion of gene pksCT in M. aurantiacus resulted in significantly decreased citrinin production. Furthermore, this ΔpksCT mutant exhibited a stronger ability to produce red and yellow pigments (Fu et al., 2007). To further investigate the mutual regulation between pigments and citrinin metabolism in M. purpureus LQ-6, the kinetics of pigments and citrinin biosynthesis were analyzed (Fig. 5C and 5D). According to the experimental results, a small amount of MPs were still detected in SBF of ΔpigR with 87.74% production reduced. However, through PCR verification and sequencing analysis (Fig. S3), the pigR gene in M. purpureus LQ-6 has indeed been completely knocked out. The pigment production after fermentation for 252 h was enhanced by 1.14-fold (from 107.10 U/mL to 121.83 U/mL, or calculated as 2945 U) after deletion of the gene pksCT, but almost no citrinin was produced. However, the concentration of citrinin was dramatically increased by 2.03-fold (from 1.85 mg/L to 3.75 mg/L, or calculated as 0.38 mg) that produced by the wild-type strain M. purpureus LQ-6 after disruption of the pigment biosynthetic pathway. These results state that different Monascus strains may exist differences after disruption of the same gene, this may be also the reason why M. purpureus ΔpigR constructed in this study still produced pigments. In addition, citrinin and MPs biosynthesis share the same starting unit (acetyl-coA) and extending unit (malonyl-coA) (Hajjaj et al., 1999). The disruption of MPs biosynthesis pathway can increase the production of citrinin, which may be due to more initial and extension units for citrinin biosynthesis. This illustrated that blocking of citrinin or pigment biosynthesis could significantly cause a change in their production in M. purpureus, and there is a mutual regulation between pigment and citrinin metabolism.
MPs biosynthesis involves the polyketide synthesis pathway and fatty acid synthesis pathway, in which many NADH/NADPH-dependent redox enzymes are involved. It’s speculated that intracellular cofactors metabolism would be disrupted after deletion of genes pigR and pksCT. Recombination strain M. purpureus ΔpigCT had the highest pigment production, because the energy used to synthesize citrinin supports the pigment biosynthesis pathway, thereby promoting disruption of the pigment production of pksCT. The kinetics analysis of pigment and citrinin biosynthesis showed logarithmic prophase of fermentation of secondary biosynthesis at 96 h (Fig. 5C and 5D). In this phase, a large amount of energy and reducing power are needed to transport the intracellular nutrients to promote the synthesis of metabolic intermediates and support cell growth (Huang et al., 2017). Additionally, the biosynthetic pathway of pigments demands more primary metabolites, such as NADH, NADPH, acetyl-CoA, and malonyl-CoA (Huang et al., 2017; Hu et al., 2012). As shown in Table 1, the level of intracellular NADH and NADPH was sharply decreased by 1.58-fold and 3.71-fold compared to that in the wild-type strain LQ-6 in recombination strain M. purpureus ΔpksCT at 4 days, respectively, and just slightly enhanced by 1.05-fold and 1.22-fold in M. purpureus ΔpigR. The concentration of NADH was almost equal between recombinants and parent strain LQ-6 at 6 days (the stationary phases of cell growth, and the logarithmic phase of pigment biosynthesis), but the concentration of NADPH was significantly reduced, especially to M. purpureus ΔpksCT (reduced by 81.97%). This indicates that deletion of gene pksCT can significantly reduce the concentration of NADH and NADPH, especially NADPH, and disrupting the gene pigR slightly affects cofactors metabolism. Based on the analysis of metabolic pathways of pigments and citrinin (Fig. 1), hexketide used in the biosynthesis of citrinin was accumulated after deletion of gene pksCT, promoting more hexketide chromophores to synthesize pigments with more NADPH and NADH consumption, resulting in low concentrations of NADH and NADPH in M. purpureus ΔpksCT at fermentation prophase (seen the data of 2 days and 4 days in Table 1), and reducing the growth rate. It may be that the reducing energy originally used for pigment synthesis in M. purpureus ΔpigR flows to the citrinin synthesis pathway, solving the feedback inhibition of pigment on growth after disruption of pigment biosynthesis. Additionally, a lot of energy, especially NADPH (directly used for pigment biosynthesis) was used for biomass synthesis pathway (such as fatty acid synthesis pathway and lipid synthesis pathway), resulting in the increased cell growth rate and an almost constant concentration of intracellular NADH and NADPH. These results suggested that cofactors are closely related to the synthesis of secondary metabolites, which provides a strategy for improving pigment production.
In this study, the levels of intracellular cofactor NADPH, NADH, glucose consumption, and cell growth were disturbed to varying degrees after deletions of gene pigR and pksCT, as well as the morphology of mycelium. There is a mutual regulation effect between the production of secondary metabolites and mycelial morphology in submerged fermentation by M. purpureus. In conclusion, this study improves our understanding of the relationship between secondary metabolites and morphology in M. purpureus and provides a strategy for maintaining high MPs production for foods and pharmaceutical fields.
References
Balakrishnan B, Karki S, Chiu SH, Kim HJ, Suh JW, Nam B, Yoon YM, Chen CC, Kwon HJ. Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 97: 6337-6345 (2013)
Chen YP, Tseng CP, Chien IL, Wang WY, Liaw LL, Yuan GF. Exploring the distribution of citrinin biosynthesis related genes among Monascus species. J. Agric. Food Chem. 56: 11767-11772 (2008)
Chen G, Bei Q, Huang T, Wu ZQ. Tracking of pigment accumulation and secretion in extractive fermentation of Monascus anka GIM 3.592. Microb. Cell Fact. 16: 172 (2017)
Chen G, Wang MH, Tian XF, Wu ZQ. Analyses of Monascus pigment secretion and cellular morphology in non-ionic surfactant micelle aqueous solution. Microb. Biotechnol. 11: 409-419 (2018)
Fu GM, Xu Y, Li YP, Tan WH. Construction of a replacement vector to disrupt pksCT gene for the mycotoxin citrinin biosynthesis in Monascus aurantiacus and maintain food red pigment production. Asia Pac. J. Clin. Nutr. 16 Suppl 1: 137-142 (2007)
Hajjaj H, Klaébé A, Loret MO, Goma G, Blanc P, Francois JM. Biosynthetic pathway of citrinin in the filamentous fungus Monascus ruber as revealed by 13C nuclear magnetic resonance. Appl. Environ. Microb. 65: 311-314 (1999)
Hu ZQ, Zhang XH, Wu ZQ, Qi HS, Wang ZL. Export of intracellular Monascus pigments by two-stage microbial fermentation in nonionic surfactant micelle aqueous solution. J. Biotechnol. 162: 202-209 (2012)
Huang T, Tan HL, Lu FJ, Chen G, Wu ZQ. Changing oxidoreduction potential to improve water-soluble yellow pigment production with Monascus ruber CGMCC 10910. Microb. Cell Fact. 16: 208 (2017)
Ibrahim D. Effect of agitation speed on the morphology of Aspergillus nigerHFD5A-1 hyphae and its pectinase production in submerged fermentation. World J. Biol. Chem. 6: 265 (2015)
Jia XQ, Xu ZN, Zhou LP, Sung CK. Elimination of the mycotoxin citrinin production in the industrial important strain Monascus purpureus SM001. Metab. Eng. 12: 1-7 (2010)
Kim D, Ku S. Beneficial effects of Monascus sp. KCCM 10093 pigments and derivatives: A mini review. Molecules. 23:98 (2018)
Kim HJ, Kim JH, Oh HJ, Shin CS. Morphology control of Monascus cells and scale-up of pigment fermentation. Process Biochem. 38: 649-655 (2002)
Kwon HJ, Balakrishnan B, Kim YK. Some Monascus purpureus genomes lack the Monacolin K biosynthesis locus. J. Appl. Biol. Chem. 59: 45-47 (2016)
Lai Y, Wang L, Qing L, Chen FS. Effects of cyclic AMP on development and secondary metabolites of Monascus ruber M-7. Lett. Appl. Microbiol. 52: 420-426 (2011)
Kim HJ, Ji GE, Lee I. Natural occurring levels of citrinin and Monacolin K in Korean Monascus fermentation products. Food Sci. Biotechnol. 16: 142-145 (2007)
Lin L, Wu SF, Li ZJ, Ren ZY, Chen MH, Wang CL. High expression level of mok E enhances the production of Monacolin K in Monascus. Food Biotechnol. 32: 35-46 (2018)
Liu J, Guo T, Luo YC, Chai XY, Wu JY, Zhao W, Jiao PF, Luo FJ, Lin QL. Enhancement of Monascus pigment productivity via a simultaneous fermentation process and separation system using immobilized-cell fermentation. Bioresour. Technol. 272: 552-560 (2019)
Lv J, Zhang BB, Liu XD, Zhang C, Chen L, Xu GR, Cheung PCK (2017) Enhanced production of natural yellow pigments from Monascus purpureus by liquid culture: The relationship between fermentation conditions and mycelial morphology. J. Biosci. Bioeng. 124: 452-458
Ning ZQ, Cui H, Xu Y, Huang ZB, Tu Z, Li YP(2017) Deleting the citrinin biosynthesis-related gene, ctnE, to greatly reduce citrinin production in Monascus aurantiacus Li AS3.4384. Int. J. Food Microbiol. 241: 325-330
Papagianni M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 22: 189-259 (2004)
Patakova P. Monascus secondary metabolites: production and biological activity. J. Ind. Microbiol. Biot. 40: 169-181 (2013)
Shao YC, Ding YD, Zhao Y, Yang S, Xie BJ, Chen FS. Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World J. Microbiol. Biotechnol. 25: 989-995 (2009)
Shao YC, Lei M, Mao ZJ, Zhou YX, Chen FS. Insights into Monascus biology at the genetic level. Appl. Microbiol. Biotechnol. 98: 3911-3922 (2014)
Shimizu T, Kinoshita H, Ishihara S, Sakai K, Nagai S, Nihira T. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl. Environ. Microbiol. 71: 3453-3457 (2005)
Sun XW, Wu HF, Zhao GH, Li ZM, Wu XH, Liu H, Zheng ZM. Morphological regulation of Aspergillus niger to improve citric acid production by chsC gene silencing. Bioproc. Biosyst. Eng. 41: 1029-1038 (2018)
Terán HR, de Souza RA, Marcelino PF, da Silva SS, Dragone G, Mussatto SI, Santos JC. Sugarcane bagasse hydrolysate as a potential feedstock for red pigment production by Monascus ruber. Food Chem. 245: 786-791 (2018)
Vecht-Lifshitz SE, Magdassi S, Braun S. Pellet formation and cellular aggregation in Streptomyces tendae. Biotechnol. Bioeng. 35: 890-896 (1990)
Wang B, Zhang XH, Wu ZQ, Wang ZL. Investigation of relationship between lipid and Monascus pigment accumulation by extractive fermentation. J. Biotechnol. 212: 167-173 (2015)
Xie NN, Liu QP, Chen FS. Deletion of pigR gene in Monascus ruber leads to loss of pigment production. Biotechnol. Lett. 35: 1425-1432 (2013)
Xie NN, Zhang YP, Chen FS. Identification of a pigment-polyketide synthase gene deleted mutant of Monascus ruber M7. Wei Sheng Wu Xue Bao. 55: 863-872 (2015)
Yang XL, Dong Y, Liu GR, Zhang C, Cao YP, Wang CT (2019) Effects of nonionic surfactants on pigment excretion and cell morphology in extractive fermentation of Monascus sp NJ1. J. Sci. Food Agric. 99: 1233-1239
Zhang C, Liang J, Yang L, Chai SY, Zhang CX, Sun BG, Wang CT. Glutamic acid promotes monacolin K production and monacolin K biosynthetic gene cluster expression in Monascus. AMB Express. 7: 22 (2017)
Zhang K, Yu C, Yang ST. Effects of soybean meal hydrolysate as the nitrogen source on seed culture morphology and fumaric acid production by Rhizopus oryzae. Process Biochem. 50: 173-179 (2015)
Acknowledgements
The authors would like to thank Prof. Tiangang Liu from Wuhan University, China, for kindly providing the pXS plasmid for cloning selection marker G418 fragment and Prof. Feng Yu from Hunan University, China, for kindly providing A. tumefaciens EHA105.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 31571874); 2011 Collaborative Innovation Center of Hunan province (2013, No. 448); Natural Science Foundation of Hunan Province (No. 2018JJ3872); Natural Science Foundation of Hunan Province (No. 2018JJ2672); Key Laboratory of Staple Grain Processing, Ministry of Agriculture and Rural Affairs of the People’s Republic of China (No. DZLS201706); Research and Development Plan in Key Areas of Hunan Province (No. 2019NK2111); and Natural Science Foundation of Hunan Province (No. 6082019JJ40542).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors of this study has any financial interest or conflict with industries or parties.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
The construction process of the pigR deletion plasmid pCAMBIA3300-ΔpigR (JPEG 99 kb)
Fig. S2
Supplementary material 2 (PDF 50 kb)
Fig. S3
Supplementary material 3 (JPEG 47 kb)
Fig. S4
Supplementary material 4 (JPEG 458 kb)
Fig. S5
Supplementary material 5 (JPEG 1927 kb)
Rights and permissions
About this article
Cite this article
Chai, X., Ai, Z., Liu, J. et al. Effects of pigment and citrinin biosynthesis on the metabolism and morphology of Monascus purpureus in submerged fermentation. Food Sci Biotechnol 29, 927–937 (2020). https://doi.org/10.1007/s10068-020-00745-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-020-00745-3