Abstract
In this study, yeasts were isolated and characterized from twelve traditional sourdough samples which belongs to Black Sea and Aegean regions of Turkey. Twenty six yeast species were isolated and identified by both 26S rDNA sequencing and FTIR spectroscopy. Saccharomyces cerevisiae (50%), Torulaspora delbrueckii (40%) and Kluyveromyces marxianus (10%) were found in 12 Turkish traditional sourdough samples. S. cerevisiae was found to be the most dominant species in Aegean region while T. delbrueckii was the most frequently isolated species in Black Sea region. Some technological properties of isolated yeast species such as acidity development, resistance to NaCI and potassium sorbate, and yeast effect on bread quality were investigated. Breads were prepared by S. cerevisiae TGM38 strain demonstrated the highest bread volume compared the other yeasts used in the study. This study showed the yeast diversity and technological properties of traditional Turkish sourdough breads fermented by chosen yeast species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Traditional sourdough breads have drawn consumer’s interest in recent years due to their nutritional, health and technological benefits. Identification and characterization of sourdough microorganisms are the first stages to understand sourdough characteristics. Studies based on the traditional sourdough microbiota constituted by yeasts and lactic acid bacteria are also increasing due to the demand for using well-characterized starter microorganisms in fermentation and product stability.
Yeasts have been used since ancient times especially in bakery and beverage products. Many valuable food ingredients and processing aids are obtained from yeast fermentations. Sourdough, one of the oldest biotechnological products, is the alternative to the baker’s yeast in the modern bakery. Specific interactions between lactic acid bacteria (LAB) and yeast take place in sourdough ecosystem. The ratio between LAB and yeast in sourdough is generally 100:1 [1]. Although, LAB is the dominant microbial group in sourdough microbiota and mainly determines the sensory, technological, nutritional, and functional features of the resulting baked products. The yeast species found in sourdough are also important for their adaptation to the specific and stressful environment created mainly by a low pH, high carbohydrate concentrations and high cell densities of LAB [2]. Yeasts contribute to the improvement of organoleptic properties and technological quality of bread by producing important metabolites during fermentation. 2,3-butanedione and phenylacetaldehyde as the most aroma active compounds were produced by the yeast metabolism, therefore honey-like and buttery caramel flavor were formed in bread. Moreover, increasing in dough volume through the production of carbon dioxide is the important function of yeasts in bread making [3, 4]. Many studies in different countries have been carried out to identify yeast species in sourdough ecosystem due to these advantages of yeasts in bakery technology. Recently, De Vuyst et al. [5] reviewed the yeast composition of 287 sourdoughs papers since 1971 and revealed the most encountered yeast species independently of identification methods. According to decreasing index of abundance, these species are: Saccharomyces cerevisiae, Candida humilis, Wickerhamomyces anomalus (syn. Pichia anomala), Torulaspora delbrueckii, Kazachstania exigua, Pichia kudriavzevii, and Candida glabrata [2].
PCR-based methods, DNA-sequencing methods and spectroscopic methods such as MALDI-ToF and FT-IR are the some examples of the techniques using in identification of microorganism in food ecosystem. Nowadays, reliable and rapid identification techniques has gained a growing interest in many research area. FT-IR spectroscopy introduced by the group of Naumann [6] is one of them. Unlike the DNA based methods, this fingerprinting technique reflects the overall chemical composition of the cells under investigation by the absorption of IR radiation. A spectral data library for yeast comprising 322 strains from 73 species was developed by Kümmerle et al. [7] with 97.5% identification accuracy in order to analyze unknown yeast species from various samples [8]. Wenning et al. [9] developed a standardized procedure for cultivation and sample preparation for the identification of yeasts. In this procedure, FT-IR microspectroscopy was used and 92% correct identification at the strain level for S. cerevisiae and 91% for D. hansenii were achieved. This kind of rapid identification technique is generally practiced in the field of clinical microbiology to characterize food-borne yeasts. On the other hand, food ecosystem such as sourdough is also required this kind of rapid identification technique in order to monitor the microbial quality, diversity, or functionality of the fermented product. The aim of this study was to characterize yeasts’ microbiota of 12 Turkish sourdough by using both 26S-rDNA gene sequencing and FT-IR spectroscopy in the HTS-XT module besides determining some technological properties and bread making performance.
Materials and methods
Materials
Twelve sourdough used in this study were collected from different artisan bakeries located in Black Sea (4 samples) region and Aegean region (6 samples) of Turkey. Two sourdough were produced in our laboratory according to an artisan bakery’s protocol. Laboratory production sourdough-1 was prepared with wheat flour while laboratory production sourdough-2 was prepared with Tarhana, spontaneously. Commercial wheat flour obtained from Istanbul Municipality Bakery, having 11.1% protein (d.b.), 0.72% ash content, and 13.8% moisture content was used for bread making.
Total titratable acidity (TTA) and pH level of sourdough samples
Ten (10) grams of sourdough were homogenized in 90 mL distilled water with a magnetic stirrer (IKA basic 2 RH, Germany), the pH value was measured by a pH meter (H35010, HANNA Instruments, Italy). For the determination of total titratable acidity (TTA), these suspension were titrated with 0.1 M NaOH to final pH 8.5. TTA was expressed as the amount of 0.1 M NaOH 10 g−1 consumed in 1 mL.
Yeast isolation from sourdough samples
Ten grams of each sample were homogenized with 90 mL of sterile peptone water (1%) for the enumeration and isolation of yeast. Yeast were counted on DRBC Agar (Merck, Germany) incubated for 24–48 h at 28 °C. Then, colonies were randomly picked from the dilution plates at higher dilution levels according to their morphology, color, and size. Yeasts were purified by the conventional streaking technique on a DRBC agar plate in order to obtain pure colonies. After incubation at 28 °C for 24–48 h, the isolates were transferred to 5 mL Malt Extract Broth (Merck, Germany). Yeasts were grown overnight in Malt Extract Broth.
DNA extraction
One milliliter of an overnight culture was centrifuged at 4 °C 13,000 g for 10 min. After the pellets were obtained, DNA extraction was performed with GF-1 nucleic acid extraction kit (Vivantis, Malaysia) according to manufacturer’s protocol.
26S rDNA sequencing analysis
DNA amplification was carried out as described by Cocolin et al. [10] with slight modification. 26S rRNA gene was amplified by PCR using the primer NL1, 5P-GCC ATA TCA ATA AGC GGA GGA AAA G-3P and a reverse primer LS2, 5P-ATT CCC AAA CAA CTC GAC TC-3P. PCR was performed in a final volume of 25 µL containing 15 µL 2X Taq Master Mix (vivantis, containing Taq DNA polymerase (0.05 U/µL), reaction buffer (2X Vibuffer A), 3.0 mM), 5 µL Nuclease free water (vivantis), 2 µL 12.5 µM of primers and 1 µL extracted DNA. The reactions were run for 30 cycles: denaturation was at 95 °C for 60 s, annealing at 52 °C for 45 s and extension at 72 °C for 60 s. An initial 5 min denaturation at 95 °C and a final 7 min extension at 72 °C were used. Products were exposed to electrophoresis in same conditions as described below. Gel was scanned on Gel documentation system (GelDoc-XR, Bio-Rad, France). The PCR product was purified with GF-1 PCR Clean up kit (Vivantis) according to manufacturer’s protocol. PCR products were sequenced by Iontek, Istanbul, Turkey. The resulting 26S rDNA sequencing were used for homology comparison using the NCBI BLAST nucleotide search database (http://blast.ncbi.nlm.nih.gov/ blast.cgi). The phylogenetic trees were constructed by using the Neighbor-Joining distance method with 2000 bootstrap replicates using PAUP version 4.0 beta 10.
FTIR spectroscopy
All spectra were determined according to Kümmerle et al. [7]. Yeast strains were cultivated on yeast extract glucose chloramphenicol agar (YGC) agar (Merck, Germany) at 27 °C for 24 h. Cells were suspended in 100 µL of distilled water. Of this suspensions 25 µL each were transferred onto the ZnSe table and were dried in an incubator at 40 °C. FTIR measurements were performed using a HTS-XT unit coupled to a Tensor 27 spectrometer (Bruker Optik GmbH, Germany) in the range of 4000–600 cm−1 wave numbers. Data were processed with the software OPUS version 7.2, for microbiological identification.
Determination of technological properties of the isolates
Properties of acidity development
The isolates inoculated in Malt extract broth for 1% (w/w) and incubated in 30 °C. pH value was measured after 3 h, 6 h, 9 h, 24 h incubation.
Determination of NaCl and potassium sorbate resistant
The isolates were grown in Malt Extract broth for two days and transferred 1 mL to 9 mL Malt Extract Broth (MEB) containing 0, 2, 4 and 6% NaCI to determine the salt resistance. Similarly, the isolates were also inoculated to MEB containing 0, 0.02, 0.05 and 0.1% potassium sorbate to determine the resistance to antifungal agent. Microbiological analyses were performed after 24 h on DRBC agar and yeast counts were enumerated after 24–48 h incubation for at 28 °C. The data were assessed as growth inhibition (%, GI):
N1, number of yeast at 0% concentrations of NaCI or potassium sorbate; N2, number of yeast at different concentrations of NaCI or potassium sorbate.
Bread production
American Association of Cereal Chemists (AACC) standard method (No 10-10) was used with some modifications according to Turkish bread formulation [11]. Control bread was prepared with 100 g flour (dry basis), 25 mL 8% yeast solution, 25 mL 6% salt solution mixed with additional water determined by Farinograph (Brabender, Germany). The yeast isolates identified as superior according to technological properties were used in formulation. For this purpose, the selected isolates were activated on Malt Extract Agar (30 °C, 24–48 h), inoculated in Malt Extract Broth (Merck, Germany) and incubated for 24 h at 30 °C. After the incubation, the isolates were centrifuged at 4 °C 13,000 g for 10 min. The pellets were used in bread dough instead of commercial yeast. All ingredients were kneaded to optimum dough development in the mixer (Kitchen aid, Model 5 K SM 150, USA), for 8 min at 4 speed. After complete mixing, dough was placed in the fermentation cabinet (Nuve TK 252, Turkey) at 30 °C and 85% relative humidity. The total fermentation time was 115 min. The baking was performed at 235 °C for 25 min in an electric oven (Maksan MKF-4P, Turkey). Afterwards, the breads were cooled to room temperature for 2 h before analyses.
Sensory evaluation
The baked breads were sliced (2 cm thickness) and sensory properties of each bread were evaluated by 20 untrained people (aged between 21 and 35 years) under normal (daylight) illumination. Descriptive profile analyses with fifteen points hedonic scale (1: dislike extremely, 2: dislike, 3: neither like nor dislike, 4: like, 5: like extremely) was used to determine the degree of overall liking of breads.
Determination of quality characteristics of breads (colour, volume and texture)
Bread samples were analyzed after cooling at room temperature for 2 h. The crust and crumb color of bread samples were measured by using Chromometer CR-100 (Konica Minolta, Japan) recording the L* (Lightness), a* (Redness to Greenness), and b* (Yellowness to Blueness) values. Bread volume was determined by applying the rapeseed displacement method according to AACC [12] with some modifications and specific volume was calculated as the volume/weight ratio (mL/g).
Texture Profile Analysis (TPA) was conducted by using TA.XT2 Plus Texture Analyzer (SMS, UK) equipped with 5 kg load cell and 36 mm diameter cylindrical compression probe. For each compression test, two slices (1.25 mm thickness of each) from the center of the bread loaf were used. The TPA method were as follows: pretest speed 1.7 mm/s; test speed 1.7 mm/s; post-test speed 1.7 mm/s; 30% compression, trigger force 5 g, the waiting time between the first and second compression cycle was 5 s.
Results and discussions
General features of sourdough samples
Total titratable acidity (TTA), pH, and the cell density of yeasts of sourdough samples were determined (data not shown). While pH of traditional sourdoughs varied between 3.75 and 4.29, titratable acidity ranged from 0.94 to 1.67% in lactic acid. The number of yeast enumerated in these samples ranged from 3.78 to 6.28 log CFU/g. Laboratory production sourdough has shown the lowest pH value (3.84) and the TTA value of this sourdough was reached up to 1.67% acidity in lactic acid. The sourdough samples which belongs to Black Sea region possessed the highest pH value (4.06 ± 0.17) and lowest TTA (1% in lactic acid), averagely. Sourdough samples which have a high pH value and low TTA shown a higher yeast cell density than the sourdough samples which have a low pH and high TTA.
The pH of sourdough is an important parameter in sourdough breadmaking due to the effect of dough reology and the activity of cereal or bacterial enzymes [13]. In this study, the pH range of 12 traditional Turkish sourdough was from 3.75 to 4.29 and the median value was 4.0. The pH range was close to 19 Italian sourdough samples (3.70–4.28) and the median value was higher than the median value of 16 French sourdough (3.50) [14, 15]. The reason of such differences is probably due to the flour characteristics using in sourdough making, especially its ash content [16]. It was also reported that pH of wheat sourdough ranged from 3.5 to 4.3 depending on the process condition and starter culture [16,17,18].
The yeast cell density in sourdough samples was between 3.78 and 6.28 log CFU/g, 5.30 ± 1.14 log CFU/g for Black Sea region, 4.96 ± 0.91 log CFU/g for Aegean region, and 4.78 ± 0.83 log CFU/g for laboratory produced sourdough. Lhomme et al. [14] found the yeast cell density from 4.70 to 7.60 log CFU/g for 16 French sourdoughs and 7.30 log CFU/g was also inditaced as a median value for 19 Italian sourdough [15]. Zhang et al. [19] revealed that the cell density of yeast isolated from 28 sourdoughs for Chinese steam bread were from 4.13 to 6.37 log CFU/g sample. From the point of yeast counts, Turkish sourdoughs in this study are closer to Chinese sourdough than the western sourdoughs. Hammes and Gänzle [20] reported that yeast counts were ranged from 5.2 to 7.1 log CFU/g in mature sourdough.
Phylogenetic tree based on 26S rDNA sequencing analysis
Isolation source of yeast strains and 26S rDNA sequencing results were shown in Table 1. T. delbrueckii was found as most dominant strains in Safranbolu sourdough from Black Sea region whereas S. cerevisiae was observed as the most dominant strains in Trabzon province of the same region. As for the Aegean region, S. cerevisiae were the dominant species in 6 sourdough samples. The sequence alignments were used to construct a phylogenetic tree by the Neighbour-Joining method (Fig. 1). 3 main group were clustered as S. cerevisiae, T. delbrueckii and K. marxianus according to Neighbour-Joining method as shown in Fig. 1. The strains of S. cerevisiae TGM 64, 33, and 62 were isolated from Black Sea region samples but they were close to the strains of S. cerevisiae isolated from Aegean region samples. This probably due to the sourdough from different region may harbor similar strains of S. cerevisiae. Lhomme et al. [14] also reported that the clustering of the strains was not related to the sourdough samples they were isolated from. The strains TGM 66, 67 and 69 were closely related to K. marxianus isolated from laboratory production sourdough-2 prepared with Tarhana.
FT-IR spectra of yeast strains
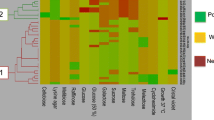
FTIR spectroscopy in combination with chemometric techniques exhibits a great potential for microbial species identification. Reliability of identification is high for both bacteria and yeasts and even sensitive typing systems obtain high classification rates [21]. In order to compare the spectra of the different strains, cluster analysis using the second derivative of the original spectra as input was carried out for different spectral regions (3030–2830, 1350–1200, 900–700 cm−1) which were worked in many previous literatures for the best discriminating features [7, 9]. These spectral windows exemplified the absorption of fatty acid compounds, carboxylic groups of proteins, RNA/DNA and phospholipid content [22, 23]. As shown in Fig. 2, 57 isolates were clustered and the dendrogram is divided in four distinct clusters. The first group of dendrogram was belonging to T. delbrueckii isolates (16 isolates), the second group includes S. cerevisiae (29 isolates) which were compose of main group and the others were S. bayanus/pasterianus (7 isolates) and K. marxianus (5 isolates). Taha et al. [24] investigated 39 yeast isolates by phenotypic, RFLP and FTIR spectroscopy method and reported that FTIR spectroscopy could be use promisingly due to its sensitivity, rapidity, high differentiation capacity and simplicity compared to conventional/molecular techniques. In our study we identified 29 yeast isolates, which identified by FTIR spectroscopy as well as partial sequencing of 26S rDNA. All yeast isolates were 96% correctly identified compared to 26S rDNA sequencing NCBI Blast search results in this study thanks to FTIR spectroscopy. Only TGM41 isolates were differentiated between two analyses. TGM41 was identified as S. cerevisiae by 26S rDNA sequencing analysis while identifying as K. marxianus by FTIR spectroscopy. S. cerevisiae was 92% correctly identified by FTIR spectroscopy identification when it compared to 26S rDNA sequence based identification. Similar result was also reported by Wenning et al. [9] for S. cerevisiae identification by FTIR spectroscopy.
Technological characterization
Acid development capabilities
Acid development properties of 20 yeast strains were investigated with 3 h intervals and 14 strains were capable of lowering the pH of sterile MEB after 24 h incubation at 30 °C below 4 (Table 2). Lower pH values are important for sourdough breadmaking in order to improve the organoleptic properties of final products. Producing organic acids by yeast in sourdough fermentation affects the flavor of the sourdough bread [25]. S. cerevisiae TGM 38 isolated from Black Sea region of Turkey showed the lowest pH value (3.73). The pH values of K. marxianus and T. delbrueckii species were between 4.18 and 4.46. In this study, the species of S. cerevisiae were more capable of lowering the pH than K. marxianus and T. delbrueckii species.
Resistance to salt and potassium sorbate
The technological characterization of isolated yeasts were evaluated under different NaCI and potassium sorbate concentration as shown in Table 2. The highest concentration of NaCI (6%) inhibited on yeast microbiota by 30–40% in 4 isolates, 20–30% in 12 isolates, 10–20% in 3 isolates. Concerning the potassium sorbate (PS) resistance of isolated yeast, 5 isolates were affected at the highest concentration of PS (1000 ppm) in the level 40–50%. The concentration of 1000 ppm PS was inhibited 30–40% in one isolate, 20–30% in 4 isolates, 10–20% in 5 isolates. T. delbrueckii TGM 19 isolated from Black Sea region/Safranbolu-1 was the most promising isolate in all thanks to showing lowest growth inhibition (GI, %) value for both NaCI and potassium sorbate conditions while T. delbrueckii TGM 15 and TGM 20 could be a good candidate for only PS conditions due to showing a GI < 10%. Similarly, S. cerevisiae TGM 41 belonging the Aegean region-1 also showed lower GI(%) value for both conditions in all S. cerevisiae species isolated in this study. K. marxianus TGM 66 was more promising candidate in all for showing the same GI (%) to NaCI and PS conditions except 1000 ppm PS. Perricone et al. [26] studied 50 yeast isolates from Almatura sourdough (Italy) to select most promising isolates as functional starter culture and found that 4 isolates showed GI > 75% and 21 isolates were partially inhibited showing GI values in the range 25–75%. Showing high values GI % under salt and potassium sorbate conditions are considerable parameter since they are generally used in bread or other cereal-based products formulation. However, yeasts belonging to the same species shows strain-specific tolerance to same conditions and also their tolerance depends on the dose and time [26, 27]. Our results also indicated that yeasts showed strain specific and dose dependent tolerance to NACI and PS.
Selection of the isolates for bread production
One T. delbrueckii and K. marxianus strain and two S. cerevisiae strains were chosen for bread producing thanks to having promising results in their technological characterization. S. cerevisiae TGM 6 and TGM 38 were chosen as a representative of two different region, Aegean region and Black Sea region, respectively (Table 1). Also, S. cerevisiae TGM 6 showed good PS resistance and S. cerevisiae TGM 38 showed the lowest pH value (Table 2) in all strains. T. delbrueckii TGM 19 were selected in bread producing because this strain showed the best NaCI (2%) resistance in 4 strains. Similarly, K. marxianus TGM 69 was selected due to its acid development capabilities (Table 3) beside being most promising candidate in terms of NaCI and PS resistance (Table 4).
Loaf volume, hardness, crust and crumb color of breads
Bread samples were produced using TGM6, TGM19, TGM38, TGM69 and commercial yeast. Quality characteristics (bread volume, specific volume, hardness, crust and crumb colour) of bread are presented in Table 3. Bread volume (mL) and specific volume (mL/g) ranged between 267.5–200 mL and 1.91–1.47 mL/g, respectively. Bread volume and specific volume of control bread produced with commercial yeast were the highest level. There was no significance differences in terms of bread volume among breads produced with TGM6, TGM19, TGM38, TGM69. Considering specific volume, the results of bread produced with TGM6, TGM19 and TGM69 were similar, otherwise TGM38 bread had higher than others except control. As shown in Table 3, the value of hardness, the peak force of the first compression of bread, were significantly different from each other. The breads produced with isolated yeast had higher hardness than the control bread. However, TGM 69 bread had the highest hardness value (14.33 N) among all samples.
Crust and crumb color of the breads were also evaluated by using a Hunterlab colorimeter (Konica Minolta, Japan). The L* (Lightness), a* (Redness to Greenness), and b* (Yellowness to Blueness) values for crust and crumb of all breads are shown in Table 3. The * values of the bread crust ranged from 59.11 (TGM69) to 54.05 (TGM19). The L* value of bread produced with K. marxianus TGM 69 (59.11) was higher than control bread (54.30) while other breads (TGM6, TGM19, TGM38) did not significantly differ from control breads in terms of L* value. The a* and b* values of bread crust were generally similar to each other. In general, no significant differences were observed between crumbs of the breads in terms of b* value. As shown in Table 3, crumb color of bread samples was generally similar to each other. A significant difference in L* value of bread crumb was observed only S. cerevisiae TGM38 bread (64.73) which was found to be darker on sensory results (Table 4). On the other hand, a* value of S. cerevisiae TGM38 was similar to the control bread. Also, b* values of bread crumb did not differ from each other significantly (P < 0.05).
After 2 h from baking, sensory analysis was carried out. Panelists were asked to evaluate crust colour, cell structure, taste, smell, chewiness and overall acceptibility of bread samples (Table 4). T. delbrueckii TGM19 was generally evaluated similar to the control bread especially in overall acceptibility. Otherwise, the bread produced with K. marxianus TGM69 was generally characterized by the lowest value of all sensory parameters, whereas the control bread had the highest sensory values. This can be due to the single effect of this strain to bread characteristic is insufficient to improve the sensorial properties. Plessas et al. [25] reported that combination of K. marxianus with L. bulgaricus or L. helveticus had a positive impact on bread flavor in contrast with K. marxianus individually.
Yeasts isolated from sourdough are important for their contribution to flavor formation and technological performance of final bread. Rapid identification techniques and single effect of sourdough yeast strain to bread quality are important parameter to select best candidates for starter culture production. This study investigated the using of FTIR spectroscopy as a rapid method by comparing 26S-rDNA gene sequencing to identify sourdough yeasts. After rapid identification of yeast isolated from sourdough using FTIR spectroscopy, the bread making performance of these strains were individually evaluated and technologically characterized. As for the bread production performance, S. cerevisiae TGM 38 isolated from Black Sea region had higher specific volume than the others beside sensory acceptability. This strain also showed high acidification ability and resistance to NaCl. Our findings demonstrated the diversity of yeast species in Turkish sourdough as well as explored their potential use in baking industries.
References
Gobbetti, M. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 7:267-274 (1998)
Huys G, Daniel HM, De Vuyst L. Taxonomy and Biodiversity of Sourdough Yeasts and Lactic Acid Bacteria. Vol. 5, pp. 105–154. In: Handbook on Sourdough Biotechnology, Springer US, Boston, MA. (2013)
Birch AN, Petersen MA, Hansen AS. The aroma profile of wheat bread crumb influenced by yeast concentration and fermentation temperature. LWT - Food Sci. Technol. 50:480–488 (2013)
Valmorri S, Tofalo R, Settanni L, Corsetti, A, Suzzi G. Yeast microbiota associated with spontaneous sourdough fermentations in the production of traditional wheat sourdough breads of the Abruzzo region (Italy). Antonie Van Leeuwenhoek. 97: 119–129 (2010)
De Vuyst L, Harth H, Van Kerrebroeck S, Leroy F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 239: 26–34 (2016)
Naumann D, Helm D, Labischinski H, Giesbrecht P. The characterization of microorganisms by Fourier-transform infrared spectroscopy (FT-IR). Pp. 43–96. In: Modern techniques for rapid microbiological analysis, VCH, New York, USA, (1991)
Kümmerle M, Scherer S, Seiler H, Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207–2214 (1998)
Wenning M, Scherer S, Naumann D. Infrared spectroscopy in the identification of microorganisms. In: Handbook of Vibrational Spectroscopy. Wiley Online Library (2008)
Wenning M, Seiler H, Scherer S. Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 68:4717–4721 (2002)
Cocolin L, Bisson LF, Mills DA. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189:81–87 (2000)
AACC. Approved Method of the AACC. Method 10-10. American Association of Cereal Chemists, St. Paul, MN, USA (1990)
AACC. Approved methods of the AACC. 10th ed. American Association of Cereal Chemists St Paul, MN. (2000)
Arendt EK, Ryan LAM, Dal Bello F. Impact of sourdough on the texture of bread. Food Microbiol. 24:165–174 (2007)
Lhomme E, Lattanzi A, Dousset X, Minervini F, De Angelis M, Lacaze G, Onno B, Gobbetti M. Lactic acid bacterium and yeast microbiotas of sixteen French traditional sourdoughs. Int. J. Food Microbiol. 215:161–70 (2015)
Minervini F, Di Cagno R, Lattanzi A, De Angelis M, Antonielli L, Cardinali G, Cappelle S, Gobbetti M. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: Interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. AEM-07721 (2012)
Collar Esteve C, Benedito de Barber C, Martínez-Anaya M. Microbial sour doughs influence acidification properties and breadmaking potential of wheat dough. Jou Food Sci. 59:629–633 (1994)
Thiele C, Gänzle MG, Vogel RV. Contribution of Sourdough Lactobacilli, Yeast, and Cereal Enzymes to the Generation of Amino Acids in Dough Relevant for Bread Flavor. Cereal Chem. 79:45–51 (2002)
Wehrle K, Arendt EK, Rheological changes in wheat sourdough during controlled and spontaneous fermentation. Cereal Chem. 75:882–886 (1998)
Zhang J, Liu W, Sun Z, Bao Q, Wang F, Yu J, Chen W, Zhang H. Diversity of lactic acid bacteria and yeasts in traditional sourdoughs collected from western region in Inner Mongolia of China. Food Control. 22:767–774 (2011)
Hammes WP, Gänzle MG. Sourdough breads and related products. In Microbiology of Fermented Foods. Springer US. pp. 199–216 (1998)
Wenning M, Scherer S. Identification of microorganisms by FTIR spectroscopy: Perspectives and limitations of the method. Appl. Microbiol. Biotechnol. 16: 7111–7120 (2013)
Naumann D. Infrared Spectroscopy in Microbiology. Encycl. Anal. Chem. 102–131 (2000)
Udelhoven T, Naumann D, Schmitt J. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 54:1471–1479 (2000)
Taha M, Hassan M, Essa S, Tartor Y. Use of Fourier transform infrared spectroscopy (FTIR) spectroscopy for rapid and accurate identification of yeasts isolated from human and animals. Int. J. Vet. Sci. Med. 1:15–20 (2013)
Plessas SA, Bekatorou J, Gallanagh P, Nigam A, Koutinas A, Psarianos C. Evolution of aroma volatiles during storage of sourdough breads made by mixed cultures of Kluyveromyces marxianus and Lactobacillus delbrueckii ssp. bulgaricus or Lactobacillus helveticus. Food Chem. 107:883–889 (2008)
Perricone M, Bevilacqua A, Corbo MR, Sinigaglia M. Technological characterization and probiotic traits of yeasts isolated from Altamura sourdough to select promising microorganisms as functional starter cultures for cereal-based products. Food Microbiol. 38:26–35 (2014)
Manini F, Casiraghi MC, Poutanen K, Brasca M, Erba D, Plumed-Ferrer C. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT - Food Sci. Technol. 66:275–83 (2016)
Acknowledgements
This study was supported by Scientific Research Project Coordination Office, Yildiz Technical University (Project Number: 2012-07-05-KAP06) and General Directorate of Agricultural Research and Policies in Turkey.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arici, M., Ozulku, G., Yildirim, R.M. et al. Biodiversity and technological properties of yeasts from Turkish sourdough. Food Sci Biotechnol 27, 499–508 (2018). https://doi.org/10.1007/s10068-017-0282-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-017-0282-0