Abstract
Objectives
The association of interleukin-6 (IL-6) -174G/C (rs1800795) and IL-6 -572G/C (rs1800796) single-nucleotide polymorphism (SNP) with the risk of acquiring rheumatoid arthritis (RA) was inconsistent among previous studies. This paper aims to investigate the association between IL-6 promoter polymorphism with RA in different ethnics.
Methods
Relevant studies were searched using Medline and Google Search engines; STATA software was used to perform the meta-analysis. Pooled odds ratios (OR) were calculated to estimate the potential genetic associations. Subgroup analysis and sensitivity analysis were applied to explore the sources of heterogeneity. Lastly, we used TSA (trial sequential analysis) software to verify the reliability of meta-analysis results.
Results
A total of 18 studies were included, involving 8116 subjects (3820 RA patients and 4296 controls). We found a tendency to associate RA with the IL-6 -174G/C allele in Asians (C vs G: OR = 4.56, 95% CI = 1.85–11.23; P < 0.001); with IL-6 -572G/C genotype or allele frequencies, there was no statistical differences between RA patients and controls (P > 0.05). TSA results indicate that the current meta-analysis can draw conclusions.
Conclusions
IL-6-174G/C gene polymorphism were associated with increased risk of RA in Asians, but not in Caucasians. There was no association between IL-6 -572G/C gene polymorphism and the risk of RA.
Key Points • Although the association between interleukin-6 (IL-6) promoter polymorphism and rheumatic arthritis (RA) has been discussed in the previous meta-analysis, their conclusions are inconsistent. • In this study, trial sequential analysis (TSA) was introduced into the meta-analysis, and the following two important conclusions were confirmed: (1) IL-6-174G/C gene polymorphism was associated with increased risk of RA in Asians, but not in Caucasians. (2) There was no association between IL-6 -572G/C gene polymorphism and the risk of RA. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a common and complex systemic autoimmune disease characterized by the chronic inflammatory synovitis and destructive changes in the bone; its multiple organ feature can lead to severe disability and even death [1]. Although the exact immunopathogenesis of arthritic inflammation is unclear, immunological dysregulation by inflammatory cytokines have been shown to play important roles in the occurrence and progression of RA [2, 3]. There was evidence that synovial cells, infiltrated monocytes/macrophages, and lymphocytes in synovial tissues of RA patients can produce a large number of cytokines and inflammatory mediators by means of autocrine or paracrine and participate in the pathogenic process of RA [4]. Interleukin-6 (IL-6) is one of these pro-inflammatory cytokines; the levels of IL-6 and soluble IL-6 receptor (sIL6R) in serum and synovial fluid of RA patients were significantly higher than those of healthy subjects [5, 6].
IL-6 comes from a wide range of sources and can be produced by various types of lymphoid and non-lymphoid cells. IL-6 gene is located in region 1 of the short arm of chromosome 7, with a total length of 5 kb, consisting of 5 exons and 4 introns, showing genetic polymorphism [7]. Recently, it has been shown that -174G/C (rs1800795) and -572G/C (rs1800796) polymorphisms of IL-6 promoter affect the expression of IL-6 [8,9,10,11]. However, these positive associations have not been consistently replicated. For example, two studies conducted in India about IL-6-174G/C failed to detect any association with RA [12, 13]. Besides, multiple studies also reported no difference in distribution between alleles and genotypes in RA patients and controls [14,15,16,17,18,19]. Interestingly, almost all studies from China have shown that C alleles could increase the susceptibility to RA [20,21,22,23,24], but You et al. did not find this conclusion [8]. On the other hand, Zhuang et al. proposed that G allele of IL-6 -572G/C may have protective effect against RA [25]; however, other studies have not drawn that conclusion [10, 14, 26]. These discrepancies could be due to the effects of genotyping method differences, publication bias, ethnic differences, and small sample size. In our study, we performed a meta-analysis to overcome the limitations of individual studies and resolve disagreements in their results.
In epidemiological studies, the size of the sample has a decisive effect on the reliability of research conclusions. As a statistical method of calculating the combined effects of multiple research subjects, a meta-analysis not only expands the sample size but also enhances the accuracy and robustness of the results. However, meta-analyses are constantly updated as new studies continue to be included. There is evidence that P < 0.05 is repeatedly used for statistical difference test; the probability of type I errors (false positives) is between 10 and 30% [27,28,29]. To minimize the increased risk of repetitive hypothesis testing due to inclusion of new research, trial sequential analysis (TSA) was introduced into this meta-analysis. With this meta-analysis, we aimed to determine the overall association of IL-6 gene rs1800795 and rs1800796 polymorphism with RA risk and to assess whether the association varies by ethnicity. We also searched the GWAS database to verify the robustness of the results.
Materials and methods
Search strategy
We performed a comprehensive search in Web of Science, Chinese Biomedical Database (CBM), Chinese National Knowledge Infrastructure (CNKI), and PubMed, using the following keywords: interleukin-6, IL-6, rheumatoid arthritis, gene, polymorphisms, and promoter. In addition, eligible publications in the bibliography list of relevant papers were evaluated. Final retrieval will take place in July 2020, without language or date constraints.
Inclusion and exclusion criteria
Included studies must meet all the following criteria: (1) case–control studies about IL-6 polymorphisms and RA in human beings; (2) sufficient genotype data presented to calculate the odds ratios (ORs) and 95% confidence intervals (CIs); and (3) full text in English or Chinese available. The exclusion criteria were as follows: (1) not a case–control design; (2) genotype frequency not reported; and (3) abstracts and reviews.
Data extraction and quality assessment
Relevant data were evaluated carefully and independently by two reviewers (Ming Shao and Huimin Xie), and disagreement was resolved with another author (Faming Pan). The relevant data were extracted: first author, publication year, country of the study population, the number of case–control samples, genotyping method, study site, genotype, and allele frequencies in each case–control study. The probability value (p value) of Hardy–Weinberg equilibrium (HWE) was also calculated. The quality of the included studies was assessed based on the Newcastle–Ottawa Scale (NOS) by two reviewers independently [30].
Statistical analyses
For included articles, the HWE of genotypes in the controls was evaluated using the Pearson chi-square test. The role of allele C in the risk of RA was targeted. Thus, an analysis was made using allelic model (C versus G), homozygote model (CC versus GG), heterozygote model (GC versus GG), dominant model (GC + CC versus GG), and recessive model (CC versus GC + GG). Between-study heterogeneities were evaluated with I2 statistic. When P < 0.1 or I2 > 50%, a high level of heterogeneity between studies was envisaged and random-effect model was adopted. Otherwise, analyses would be conducted with fixed-effect models (Mantel- Haenszel method). We also conducted subgroup analyses by ethnicity of participants. The effect of IL-6 -174G/C and IL-6 -572G/C on the risk of RA was measured by P value, odds ratio (OR), and 95% confidence interval (CI). P < 0.05 was seen as statistically significant. Ethnicity was independently determined by the two authors based on historical data combined with the original study area. Meanwhile, sensitivity analysis was conducted by repeating analysis after omitting one study each time to estimate the effect of studies quality on the final result. Publication bias was measured using Begg’s test and Egger’s tests. All statistical analyses were carried out using STATA software (STATA 11.0, StataCorp, College Station, TX, USA).
Trial sequential analysis
Due to the increased risk of random errors and repeated significance tests, meta-analysis may be affected by type I errors. Trial sequential analysis (TSA) was a useful tool to verify the reliability of the results from meta-analysis by estimating the required information size (RIS) (sample size of included studies) and calculating the threshold for statistical significance. The red curves represent the O’Brien-Fleming boundary and futility boundary. The blue line represents the cumulative Z-curve. RIS represents required information size. If the Z-curve crossed RIS line, the result of meta-analysis would be conclusive. Moreover, if the Z-curve crossed the O’Brien-Fleming boundary or futility boundary, the conclusions could also be made even before it crossed the RIS. However, if the cumulative Z value does not cross the O’Brien-Fleming boundary or futility boundary or the RIS threshold, it means the sample size is not sufficient [31], and more studies are needed to confirm the result. TSA would be conducted in allelic model in our study. Our study set the relative ratio reduction (RRR) to 20%, the first type of error α = 0.05, the second type of error β = 0.2, and control event proportion was an average of each included study to evaluate RIS. TSA was conducted on TSA version 0.9.5.10 beta (Copenhagen Trial Unit, Centre for Clinical Intervention Research, Copenhagen) [32].
Results
Characteristics of the included studies
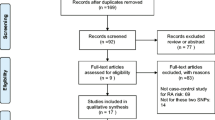
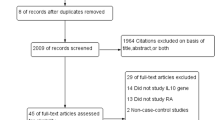
The selection process of literature retrieval is shown in Fig. 1. To put it simply, we found 2713 references through electronic databases and other channels, and 2598 references were not associated with cytokine polymorphism in terms of titles and abstracts. Of the remaining 115 studies, 90 were excluded for not performing the IL-6 promoter polymorphisms. After full text evaluation, we excluded 7 articles for the following reasons: 3 were not case–control studies, 2 were review articles, and 2 were repeated data sets. Finally, 18 studies met the inclusion criteria including 3820 cases and 4296 controls. Table 1 showed the main characteristics of included studies. These 18 studies were conducted in 11 countries (number of studies): China (6); India (2); Iraq (1); Poland (2); Mexico (1); Turkey (1); the Netherlands (1); the UK (1); Macedonia (1); Egypt (1); and Spain (1). There were 4 studies in which the genotype distribution of the control group did not conform to HWE (P < 0.05) [10, 22, 23, 26]. The NOS results showed that these studies had scores ranging from 6 to 8, with an average score of 7.56. Therefore, the methodological quality of the selected studies is generally reliable. Table 1 shows that the inter-ethnic frequency of the major allele -174G has been found to vary enormously.
Meta-analysis results
The association and heterogeneity test results between IL-6 promoter polymorphism and RA are shown in Table 2. Overall, a significant association of IL-6 -174G/C polymorphism with an increased risk of RA was observed in the allelic model (C vs G: OR = 1.83, 95% CI = 1.31–2.54; P < 0.001), heterozygote model (GC vs GG: OR = 1.83, 95% CI = 1.20–2.79; P = 0.005), and dominant model (CC + GC vs GG: OR = 2.23, 95% CI = 1.51–3.29; P < 0.001), but not in the homozygote recessive models. The relationship between IL-6 -174G/C SNP and RA risk was also analysed by subgroup analysis of ethnic stratification. The result of the subgroup analysis was very interesting. In Caucasians, none of the models showed a significant association between IL-6 -174G /C polymorphism and RA risk. In contrast, IL-6 rs1800795 was significantly associated with increased RA risk in Asians. The pooled OR and 95% CI for the various genetic models were: allelic (C vs G: OR = 4.56, 95% CI = 1.85–11.23, P = 0.001); homozygous (CC vs GG: OR = 5.09, 95% CI = 2.35–11.04, P < 0.001); heterozygous (GC vs GG: OR = 4.59, 95% CI = 1.71–12.31, P = 0.001); dominant (GC + CC vs GG: OR = 5.27, 95% CI = 1.96–14.11, P < 0.001); and recessive (CC versus GC + GG: OR = 4.06, 95%CI = 1.94–8.481, P < 0.001). There was no statistical difference in IL-6 -572G/C polymorphism between cases and controls (Fig. 2).
TSA
To assess the risk of random errors, we performed a sequential analysis of the trials. The red vertical bars represent the amount of information required (sample size). Figure 3A and Fig. 3B showed that the cumulative Z-curve crossed the traditional boundary and the O’Brien-Fleming boundary, and Fig. 3C showed crossed the futility boundary and the RIS, suggesting there was no need for more evidence to establish an additional study of rs1800795 in RA. For IL-6 rs1800796, TSA studies showed that although the Z-curve did not cross the traditional boundary, it crossed the invalid boundary, indicating that this result would not change after more studies were included (Fig. 4). Perhaps there is really no correlation between RS1800796 polymorphism and RA.
Heterogeneity and publication bias diagnostics
We found significant heterogeneity in the interleukin-6 promoter polymorphism and RA-related information. Due to the large heterogeneity among studies, a single study included in the meta-analysis was omitted continuously, and the source was found through sensitivity analysis (Supplementary Fig. 1). Although we did not find the possible source of heterogeneity through sensitivity analysis, we searched the GWAS catalog (https://www.ebi.ac.uk/gwas/). It is a pity that rs1800795 was not found, but the results of GWAS on rs1800796 were consistent with our results (Supplementary Fig. 2). The findings indicated that no individual study influenced the overall. Begg’s and Egger’s tests did not suggest significant publication bias ( P > 0.05).
Discussion
RA is a systemic autoimmune disease with progressive and destructive osteoarthropathy, and its pathogenesis is complex and unclear. Previous studies of IL-6 promoter polymorphism in RA have yielded controversial results, which is not surprising as inconsistent results are common in genetic studies of complex diseases. The most likely explanations are true genetic heterogeneity, ethnic differences, clinical heterogeneity, and small sample size. As a method, meta-analysis can obtain reliable results by expanding the sample size. However, previous meta-analysis results on the correlation between IL-6 promoter polymorphisms and RA were inconsistent [13, 33,34,35]. Therefore, it is useful to adopt a new method to help prove the stability of meta-analysis results. Compared with the previous meta-analysis of this topic, our study introduced the TSA method for the first time, which further proved the reliability of the current meta-analysis results. As a multifunctional cytokine, IL-6 is involved in inflammatory and immune responses. IL-6 is located on chromosome 7p21, and the expression of IL-6 can be regulated at the transcriptional level by -174G/C and -572G/C at the 5′ end [11, 36]. IL-6 promotes the proliferation and secretion of T and B lymphocytes and causes inflammatory secretion in the acute stage, leading to cartilage and bone damage in RA patients [37]. Regarding IL-6 -174G/C gene polymorphism, our meta-analysis revealed that C alleles was associated with RA risk (Fig. 2A) and increased the susceptibility of RA in the study. When the analysis was stratified by ethnicity, we found a statistically significant relationship between rs1800795 and the increased RA risk in the Asian population under all genetic models, but not in Caucasians. As far as we know, the inter-ethnic frequency of the minor allele -174C has been found to vary enormously. The frequency of this allele has been reported to be higher in Europeans (50%), but lower in the Afro-Caribbeans (5%) [38]. There is growing evidence that some ethnically specific polymorphisms are functional, suggesting that they may play an important role in disease susceptibility. Thus, this finding suggests that the association between RA and IL-6 -174G/C polymorphism varies among ethnic groups due to the frequency of their genotypes [39]. In addition, the regional differences between Europe and Asia could be a consequence of referral bias with more severe patients being referred to hospitals. The meta-analysis results of Dar et al. also reached a consistent conclusion [13], but the meta-analysis results of Lee et al. showed that the IL-6 -174G/C polymorphism may confer susceptibility to RA in Caucasians [33]. In our study, TSA analysis results show that the Z-curve intersects the TSA boundary, indicating that the current study has been able to draw reliable conclusions.
With respect to IL-6 -572G/C gene polymorphism, our study identified that no significant differences were observed in the genotypes and alleles frequencies (Fig. 2B) between RA and controls. According to the TSA results, we believe that there is no correlation between rs1800796 alleles and RA. However, the meta-analysis of Zhang et al. showed that the IL-6 -572G/C gene polymorphism was associated with the risk of RA [34]. This is obviously contrary to the conclusion of our meta-analysis. In addition, the results of the two existing meta-analyses were consistent with our conclusion [33, 35]. On the other hand, although there may be different definitions of ethnicity, the results of Pacheco-Soto et al. support our conclusions [40]. Besides, all studies have shown no association between the IL-6 -572G/C gene polymorphism and RA susceptibility in Caucasians [9, 10, 14, 26]. However, Huang et al. and Lu et al. found that the rs1800796 C allele increased the risk of RA in Asians [20, 24]. Therefore, we believe that there is no correlation between IL-6 -572G/C gene polymorphism and RA in Caucasians, and there is no need to conduct redundant research in the future. TSA has shown that the results of the current study can be concluded. The results of the GWAS study were consistent [41].
However, there are some limitations to our study. First, the subgroup analysis was based on ethnicity, not other variables. There has been a lot of debate about the definition of ethnicity, so that might affect the analysis. In addition, it is well known that certain gene frequencies vary in disease duration, age, disease activity, and so on. Second, this meta-analysis found significant heterogeneity between studies, and possible causes of heterogeneity, such as lifestyle and environmental exposure, were not examined, which may have distorted the analysis. Third, although TSA can effectively support the conclusions in our meta-analysis, TSA will increase the risk of false negative (type II error) while strictly controlling false positive error (type I error). In other words, IL-6 -572G/C gene polymorphism may influence the RA susceptibility, but there is no such relationship in our analysis.
Conclusion
In conclusion, this meta-analysis suggests that IL-6 -174G/C gene polymorphism were associated with increased risk of rheumatic arthritis in Asians, but not in Caucasians. There was no correlation between IL-6 -572G/C gene polymorphism and the risk of RA.
References
McInnes IB, Schett G (2011) The pathogenesis of rheumatoid arthritis. N Engl J Med 365(23):2205–2219
Jančić I, Arsenović-Ranin N, Sefik-Bukilica M, Zivojinović S, Damjanov N, Spasovski V, Srzentić S, Stanković B, Pavlović S (2013) 174G/C interleukin-6 gene promoter polymorphism predicts therapeutic response to etanercept in rheumatoid arthritis. Rheumatol Int 33(6):1481–1486
Feldmann M, Brennan FM, Maini RN (1996) Role of cytokines in rheumatoid arthritis. Annu Rev Immunol 14:397–440
Miossec P (2004) An update on the cytokine network in rheumatoid arthritis. Curr Opin Rheumatol 16(3):218–222
Hirano T, Matsuda T, Turner M, Miyasaka N, Buchan G, Tang B, Sato K, Shimizu M, Maini R, Feldmann M et al (1988) Excessive production of interleukin 6/B cell stimulatory factor-2 in rheumatoid arthritis. Eur J Immunol 18(11):1797–1801
Nishimoto N, Kishimoto T (2006) Interleukin 6: from bench to bedside. Nat Clin Pract Rheumatol 2(11):619–626
Ji YF, Jiang X, Li W, Ge X (2019) Impact of interleukin-6 gene polymorphisms and its interaction with obesity on osteoporosis risk in Chinese postmenopausal women. Environ Health Prev Med 24(1):48
You CG, Li XJ, Li YM, Wang LP, Li FF, Guo XL, Gao LN (2013) Association analysis of single nucleotide polymorphisms of proinflammatory cytokine and their receptors genes with rheumatoid arthritis in northwest Chinese Han population. Cytokine 61(1):133–138
Zavaleta-Muñiz SA, Martín-Márquez BT, Gonzalez-Lopez L, Gonzalez-Montoya NG, Díaz-Toscano ML, Ponce-Guarneros JM, Ruiz-Padilla AJ, Vázquez-Del Mercado M, Maldonado-González M, Fafutis-Morris M, Flores-Martínez SE, Martínez-García EA, Gamez-Nava JI (2013) The -174G/C and -572G/C interleukin 6 promoter gene polymorphisms in mexican patients with rheumatoid arthritis: a case-control study. Clin Dev Immunol. 2013:959084
Wielińska J, Dratwa M, Świerkot J, Korman L, Iwaszko M, Wysoczańska B, Bogunia-Kubik K (2018) Interleukin 6 gene polymorphism is associated with protein serum level and disease activity in Polish patients with rheumatoid arthritis. HLA 92(Suppl 2):38–41
Arman A, Coker A, Sarioz O, Inanc N, Direskeneli H (2012) Lack of association between IL-6 gene polymorphisms and rheumatoid arthritis in Turkish population. Rheumatol Int 32(7):2199–2201
Shafia S, Dilafroze, Sofi FA, Rasool R, Javeed S, Shah ZA (2014) Rheumatoid arthritis and genetic variations in cytokine genes: a population-based study in Kashmir Valley. Immunol Invest. 43(4):349–59
Dar SA, Haque S, Mandal RK, Singh T, Wahid M, Jawed A, Panda AK, Akhter N, Lohani M, Areeshi MY, Rai G, Datt S, Bhattacharya SN, Ramachandran VG, Das S (2017) Interleukin-6-174G > C (rs1800795) polymorphism distribution and its association with rheumatoid arthritis: a case-control study and meta-analysis. Autoimmunity 50(3):158–169
Amr K, El-Awady R, Raslan H (2016) Assessment of the -174G/C (rs1800795) and -572G/C (rs1800796) Interleukin 6 gene polymorphisms in Egyptian patients with rheumatoid arthritis. Open Access Maced J Med Sci 4(4):574–577
Emonts M, Hazes MJ, Houwing-Duistermaat JJ, van der Gaast-de Jongh CE, de Vogel L, Han HK, Wouters JM, Laman JD, Dolhain RJ (2011) Polymorphisms in genes controlling inflammation and tissue repair in rheumatoid arthritis: a case control study. BMC Med Genet 12:36
Panoulas VF, Douglas KM, Smith JP, Stavropoulos-Kalinoglou A, Metsios GS, Nightingale P, Kitas GD (2009) Transforming growth factor-beta1 869T/C, but not interleukin-6 -174G/C, polymorphism associates with hypertension in rheumatoid arthritis. Rheumatology (Oxford) 48(2):113–118
Trajkov D, Mishevska-Perchinkova S, Karadzova-Stojanoska A, Petlichkovski A, Strezova A, Spiroski M (2009) Association of 22 cytokine gene polymorphisms with rheumatoid arthritis in population of ethnic Macedonians. Clin Rheumatol 28(11):1291–1300
Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, Miranda-Filloy JA, Llorca J, Martin J, Gonzalez-Gay MA (2009) Interleukin-6 gene -174 promoter polymorphism is associated with endothelial dysfunction but not with disease susceptibility in patients with rheumatoid arthritis. Clin Exp Rheumatol 27(6):964–970
Pawlik A, Wrzesniewska J, Florczak M, Gawronska-Szklarz B, Herczynska M (2005) IL-6 promoter polymorphism in patients with rheumatoid arthritis. Scand J Rheumatol 34(2):109–113
Huang XZ, Zhuang JH, Ren YG, Zhou LJ, Zhou Q (2007) Association of interleukin-6 and interleukin-18 gene polymorphism with rheumatoid arthritis in Guangdong Han population. Journal of Southern Medical University 11:1661–1664
Li YH, Sun W, SQ Wang, Yang SC (2009) Study on interleukin-6 gene promoter polymorphism in patients with rheumatoid arthritis. Clinical Focus (19): 1715–1717
Li X, Chai W, Ni M, Xu M, Lian Z, Shi L, Bai Y, Wang Y (2014) The effects of gene polymorphisms in interleukin-4 and interleukin-6 on the susceptibility of rheumatoid arthritis in a Chinese population. Biomed Res Int 265435
Li F, Xu J, Zheng J, Sokolove J, Zhu K, Zhang Y, Sun H, Evangelou E, Pan Z (2014) Association between interleukin-6 gene polymorphisms and rheumatoid arthritis in Chinese Han population: a case-control study and a meta-analysis. Sci Rep 4:5714
Lu YL,Yu XD (2009) Study on serum IL-6 level and gene polymorphism in patients with rheumatoid arthritis.Chinese Journal of Cellular and Molecular Immunology (08):725–726+728.
Zhuang JH, Ren YG, Huamg XZ, Ding HM, Zhou Q (2007) Study on the gene polymorphism of interleukin-6 promote rinrheumatoid arthritis patients of Chinese Han population in the region of Guangdong. Laboratory Medicine 06:697–702
Ad’hiah AH, Mahmood AS, Al-kazaz A, Mayouf K (2018) Gene expression and six single nucleotide polymorphisms of interleukin-6 in rheumatoid arthritis: a case-control study in Iraqi patients. Alexandria Journal of Medicine (54): 639–645
Hu M, Cappeleri J, Lan KK (2007) Applying the law of the iterated logarithm to control type in cumulative meta-analysis of binary outcomes. Clin Trials 4:329–340
Borm GF, Donders AR (2009) Updating meta-analyses leads to larger type I errors than publication bias. J Clin Epidemiol 62(8):825–830
Berkey CS, Mosteller F, Lau J, Antman EM (1996) Uncertainty of the time of first significance in random effects cumulative meta-analysis. Control Clin Trials 17(5):357–371
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25:603–605
Wetterslev J, Jakobsen JC, Gluud C (2017) Trial Sequential Analysis in systematic. reviews with meta-analysis. BMC Med Res Methodol 17(1):39
Thorlund K, Engstrøm J, Wetterslev J, et al. User manual for trial sequential analysis (TSA). Copenhagen Trial Unit. Accessed 2011. Available from: http://www.ctu.dk/tsa/files
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2012 Jul) The association between interleukin-6 polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm Res 61(7):665–671
Zhang C, Jiao S, Li T, Zhao L, Chen H (2020) Relationship between polymorphisms in -572G/C interleukin 6 promoter gene polymorphisms (rs1800796) and risk of rheumatoid arthritis: a meta-analysis. Int J Rheum Dis. 23(1):47–54
Li T, Yang L, Wang ZQ, Fu SJ (2014) Systematic evaluation on association between interleukin-6 gene polymorphism and rheumatoid arthritis susceptibility. Chongqing Medicine 43(36):4887–4890
Morse HR, Olomolaiye OO, Wood NA, Keen LJ, Bidwell JL (1999 Oct) Induced heteroduplex genotyping of TNF-alpha, IL-1beta, IL-6 and IL-10 polymorphisms associated with transcriptional regulation. Cytokine 11(10):789–795
Srirangan S, Choy EH (2010 Oct) The role of interleukin 6 in the pathophysiology of rheumatoid arthritis. Ther Adv Musculoskelet Dis 2(5):247–256
Sie MP, Sayed-Tabatabaei FA, Oei HH, Uitterlinden AG, Pols HA, Hofman A, van Duijn CM, Witteman JC (2006 Jan) Interleukin 6–174 g/c promoter polymorphism and risk of coronary heart disease: results from the Rotterdam study and a meta-analysis. Arterioscler Thromb Vasc Biol 26(1):212–217
Jeon JY, Kim HA, Kim SH, Park HS, Suh CH (2010 Nov) Interleukin 6 gene polymorphisms are associated with systemic lupus erythematosus in Koreans. J Rheumatol 37(11):2251–2258
Pacheco-Soto BT, Porchia LM, Lara-Vazquez WC, Torres-Rasgado E, Perez-Fuentes R, Gonzalez-Mejia ME (2020) The association between interleukin-6 promoter polymorphisms and rheumatoid arthritis by ethnicity: a meta-analysis of 33 studies. Reumatol Clin. S1699–258X(20):30079–6
Okada Y, Wu D, Trynka G et al (2014) Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506(7488):376–381
Magyari L, Varszegi D, Kovesdi E, Sarlos P, Farago B, Javorhazy A, Sumegi K, Banfai Z, Melegh B (2014) Interleukins and interleukin receptors in rheumatoid arthritis: Research, diagnostics and clinical implications. World J Orthop 5(4):516–536
Funding
This study was supported by grants from the National Natural Science Foundation of China (81273169, 81573218, 81773514, 82073655) and the funds for academic and technical leaders in Anhui province (2017D140).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
We have sought approval from the biomedical ethics committee of Anhui Medical University.
Conflict of interest
Ming Shao, Huimin Xie, Hui Yang, Wei Xu, Yuting Chen, Xing Gao, Shiyang Guan, Shengqian Xu, Zongwen Shuai and Faming Pan declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Supplementary Figure 1
Sensitivity analysis about the IL-6 promoter polymorphism and the risk of RA in the allele model.A) rs1800795; B) rs1800796; (JPG 264 KB)

Supplementary Figure 2
GWAS results and Meta-analysis results of the association between IL-6 promoter polymorphism and RA susceptibility. (JPG 158 KB)
Rights and permissions
About this article
Cite this article
Shao, M., Xie, H., Yang, H. et al. Association of interleukin-6 promoter polymorphism with rheumatoid arthritis: a meta-analysis with trial sequential analysis. Clin Rheumatol 41, 411–419 (2022). https://doi.org/10.1007/s10067-021-05886-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05886-2