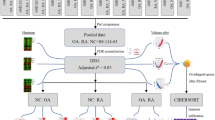
Abstract
Introduction/objectives
Rheumatoid arthritis (RA) and osteoarthritis (OA) are two common joint diseases with similar clinical manifestations. Our study aimed to identify differential gene biomarkers in the synovial tissue between RA and OA using bioinformatics analysis and validation.
Method
GSE36700, GSE1919, GSE12021, GSE55235, GSE55584, and GSE55457 datasets were downloaded from the Gene Expression Omnibus database. A total of 57 RA samples and 46 OA samples were included. The differentially expressed genes (DEGs) were identified. The Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were also performed. Protein–protein interaction (PPI) network of DEGs and the hub genes were constructed and visualized via Search Tool for the Retrieval of Interacting Genes/Proteins, Cytoscape, and R. Selected hub genes were validated via reverse transcription-polymerase chain reaction.
Results
A total of 41 DEGs were identified. GO functional enrichment analysis showed that DEGs were enriched in immune response, signal transduction, regulation of immune response for biological process, in plasma membrane and extracellular region for cell component, and antigen binding and serine-type endopeptidase activity for molecular function. KEGG pathway analysis showed that DEGs were enriched in cytokine-cytokine receptor interaction and chemokine signaling pathway. PPI network analysis established 70 nodes and 120 edges and 15 hub genes were identified. The expression of CXCL13, CXCL10, and ADIPOQ was statistically different between RA and OA synovial tissue.
Conclusion
Differential expression of CXCL13, CXCL10, and ADIPOQ between RA and OA synovial tissue may provide new insights for understanding the RA development and difference between RA and OA.
Key Points • Bioinformatics analysis was used to identify the differentially expressed genes in the synovial tissue between rheumatoid arthritis and osteoarthritis. • CXCL13, CXCL10, and ADIPOQ might provide new insight for understanding the differences between RA and OA. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is an autoimmune inflammatory disease characterized by synovial inflammation and hyperplasia, as well as cartilage and bone destruction, affecting approximately 0.5–1.0% of the world’s population. RA occurs in women more than men and mainly diagnosed in people aged 40–60. With the development of rheumatoid arthritis, joint damage, disability, and cardiovascular and other comorbidities occur [1, 2]. Genetics, epigenetics, smoking, sex, dust inhalation, gut microbiota, and modifiable lifestyle factors are risk factors in the development of RA. Genetic factors account for 50% of risk factors, thought to be specifically associated with either ACPA-positive or ACPA-negative disease [2, 3]. Due to the influence of genetic factors, RA is considered to be a heterogeneous disease. Osteoarthritis (OA) is the most common arthritis characterized by joint pain and dysfunction caused by joint degeneration and the prevalence of OA increases with age. Due to the complexity of the etiology, OA can be divided into primary or secondary OA [4]. Systemic factors (such as genetics, dietary intake, estrogen, and bone density) and biomechanical factors (such as muscle weakness, obesity, and joint laxity) are risk factors in the development of OA [5]. More studies have shown that synovial inflammation is also one of the important pathological changes in OA. The RA and OA have certain similarities in risk factors, pathological changes, and clinical manifestation; however, further studies are needed to identify the differences.
Bioinformatics is a new area in molecular biology and information technology. With the development of gene chips and next-generation sequencing technology, the field of bioinformatics is becoming more popular in the field of human genetics [6, 7]. Bioinformatics analysis is used to study a variety of diseases [8,9,10].
Here, we aimed to use bioinformatics to analyze the gene expression differences between RA and OA synovial tissue. We downloaded GSE36700, GSE1919, GSE12021, GSE55235, GSE55584, and GSE55457 datasets from the Gene Expression Omnibus (GEO) database. The Gene Ontology (GO) functional enrichment analysis, Kyoto Encyclopedia Genes and Genomes (KEGG) pathway analysis, and protein–protein interaction (PPI) network analysis were used to further analyze the differentially expressed genes (DEGs). The DEGs were further verified using reverse transcription-polymerase chain reaction (qRT-PCR) to promote a further understanding of the difference between RA and OA synovial tissue.
Materials and methods
Microarray data
Gene expression profiles datasets, GSE36700 [11], GSE1919 [12], GSE12021 [13], GSE55235 [14], GSE55584 [14], and GSE55457 [14], were downloaded from the GEO database. The GSE55235 dataset contained 30 samples; 10 RA and 10 OA synovial tissue samples, which was based on the GPL91 [HG-U133A] Affymetrix Human Genome U133A platform. The GSE1919 dataset contained 15 samples; 5 RA and 5 OA synovial tissue samples, which was based on the GPL91 [HG_U95A] Affymetrix Human Genome U95A Array. The GSE36700 dataset contained 25 samples; 7 RA and 5 OA synovial tissue samples, which was based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. The GSE12021 dataset contained 31 samples; 12 RA and 10 OA synovial tissue samples, which was based on the GPL96 [HG-U133A] Affymetrix Human Genome U133A Array. The GSE55584 dataset contained 16 samples; 10 RA and 6 OA synovial tissue samples, which was based on the GPL96 [HG-U133A] Affymetrix Human Genome U133A Array. The GSE55457 dataset contained 33 samples; 13 RA and 10 OA synovial tissue samples, which was based on the GPL96 [HG-U133A] Affymetrix Human Genome U133A Array. The Series Matrix File of GSE36700, GSE1919, GSE12021, GSE55235, GSE55584, and GSE55457 was also downloaded from the GEO database.
Identification of DEGs
The DEGs between RA and OA samples were identified using the limma package of R software [15]. P value < 0.05 and |log FC|≥ 2 were chosen as the cut-off criteria. The upregulated and downregulated genes were visualized using R 4.0.0 software.
GO functional enrichment and KEGG pathway analysis of DEGs
GO functional enrichment analysis was used to annotate genes, including biological process (BP), molecular function (MF), and cellular component (CC) [16]. GO and KEGG [17] pathways of DEGs were identified and visualized with clusterProfiler [18], ggplot2 [19] package of R. P < 0.05 was chosen as the cut-off criteria, indicating a statistically significant difference.
Integration of the PPI network and hub gene analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) [20] was used for evaluating and integrating PPI information of DEGs. The cytoHubba plugin in Cytoscape version 3.7.2 was used to screen and visualize 15 hub genes (MCC method) [21].
Total RNA extraction and qRT-PCR validation
The synovial tissue from nine patients with RA and nine with OA was harvested for qRT-PCR validation to confirm the genes identified via bioinformatics analysis. The characteristics of patients was shown in Supplementary Table 1. Total RNA was extracted from synovial tissue using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. Isolated total RNA was reverse transcribed into cDNA with PrimeScript™ RT Master Mix (Perfect Real Time) (Takara, China), and qRT-PCR was carried out using TB Green® Premix Ex Taq™ (Tli RNaseH Plus) (Takara, China). β-Actin was used as an internal reference. The relative mRNA expression was calculated using the 2−ΔΔCt method. The t-test was used for the statistical analysis, and P < 0.05 indicated a significant difference.
Results
Identification of DEGs
A total 41 DEGs, including 28 upregulated and 13 downregulated genes, were identified in our study after bath correction and standardization using R. The results were visualized with a volcano plot using the R software (Fig. 1).
GO functional enrichment and KEGG pathway analysis of DEGs
Analysis results showed that for BP, the DEGs were enriched in humoral immune response, antimicrobial humoral response, complement activation, classical pathway, positive regulation of lymphocyte activation, and lymphocyte-mediated immunity. For CC, the DEGs were enriched in immunoglobulin complex, circulating, immunoglobulin complex, blood microparticle, and external side of the plasma membrane. For MF, the DEGs were enriched in antigen binding, CXCR chemokine receptor binding, immunoglobulin receptor binding, chemokine activity, glycosaminoglycan binding, cytokine activity, and chemokine receptor binding (Fig. 2, Table 1). The significantly enriched pathway of the DEGs included cytokine-cytokine receptor interaction, PPAR signaling pathway, chemokine signaling pathway, intestinal immune network for IgA production, and toll-like receptor signaling pathway (Fig. 3, Table 2).
PPI network construction and hub gene selection in the PPI network
With the STRING database, the PPI network of DEGs was established, consisting of 36 nodes and 32 edges and the PPI enrichment P-value was 4.63 × 10−14. The cytoHubba plugin in Cytoscape was used to identify and visualize 15 hub genes: CXCL10, CXCL9, CXCL11, CXCL13, GZMK, IGJ, POU2AF1, LCK, ADIPOQ, IGLL5, IL7R, TNFRSF17, PLIN1, PCK1, and MMP1 (Fig. 4).
qRT-PCR validation of the hub genes
According to the literature analysis, we examined the relative expression of CXCL10, CXCL11, CXCL13, GZMK, LCK, ADIPOQ, and PCK1 in 9 OA and 9 RA synovial tissue via qRT-PCR. Compared to the OA synovial tissue, the expression of ADIPOQ was significantly downregulated and the expression of CXCL10 and CXCL13 were upregulated in RA synovial tissue (Fig. 5). The target gene primers are shown in Table 3.
Discussion
In this study, we screened a total of 57 RA and 46 OA synovial tissue using bioinformatics analysis. The results of the GO functional enrichment analysis, KEGG pathway analysis, the PPI network construction, and the hub gene analysis provided new insights into the molecular mechanisms between RA and OA. The occurrence of RA and OA is a complex biological process, including the abnormally expressed genes and the activation of disease-associated pathways. It is important to identify the genes and signaling pathways involved in the pathogenesis of RA and OA. The identification of the DEGs and signaling pathways of RA and OA could help the further understanding of the difference between RA and OA.
The GO functional enrichment analysis showed that the DEGs between RA and OA synovial tissue were mainly enriched in the humoral immune response, antimicrobial humoral response, lymphocyte-mediated immunity, and positive regulation of cell activation for BP; in immunoglobulin complex, circulating, external side of plasma membrane, MHC class II protein complex, and MHC protein complex for CC; and in antigen binding, CXCR chemokine receptor binding, immunoglobulin receptor binding, chemokine activity, cytokine activity, and chemokine receptor binding for MF. These functional enrichment analyses indicated that the DEGs were enriched in inflammation and immune response, which play an important role in the pathogenesis of RA [22, 23]. KEGG pathway analysis showed that the DEGs were enriched in cytokine-cytokine receptor interaction, PPAR signaling pathway, chemokine signaling pathway, rheumatoid arthritis, and toll-like receptor signaling pathway which are closely related to inflammation and immune diseases [24, 25]. These results illustrate the important role of the DEGs between RA and OA and further study is needed to confirm the conclusion.
The DEGs were further analyzed by constructing a PPI network and screening the hub genes, then we verified the selected hub genes via qRT-PCR. Expressions of CXCL10 and CXCL13 were upregulated in RA synovial tissue. The expression of ADIPOQ was downregulated in RA synovial tissue. CXCL10 is located on chromosome 4q21.1 and encodes a chemokine of the CXC subfamily and ligand for the receptor CXCR3. CXCL10 has been detected in many autoimmune diseases, such as type I diabetes, systemic lupus erythematosus, and RA. CXCL10 has been detected in the sera, synovial fluid, and synovial tissue of patients with RA [26, 27]. In the present study, we found that CXCL10 was upregulated in RA synovial tissue compared with OA synovial tissue. CXCL13 is located on chromosome 4q21.1. The synovial CXCL13 is a marker of the severe pattern of RA [28]. In our study, the expression of CXCL13 was upregulated in RA synovial tissues compared with that in OA synovial tissue. Our result was consistent with previous studies. ADIPOQ is an adipokine released from adipose tissue and plays an important role in bone formation and bone resorption [29]. ADIPOQ is located on chromosome 3q27.3 and mutations on this gene are associated with adiponectin deficiency. Adiponectin expression levels were reported to be higher in patients with OA than in the healthy controls. The expression of this gene is also increased in knee osteoarthritis synovial tissues [30], and the SNP, rs182052, of ADIPOQ is associated with susceptibility to knee osteoarthritis in the Chinese population [31], and the SNP, rs1501299, may be associated with the development of OA as previously reported [32]. In the present study, ADIPOQ was downregulated in RA synovial tissue compared with OA. It may be due to the fact that obesity is more correlated with the occurrence of OA than RA.
In conclusion, we integrated bioinformatics analysis and qRT-PCR validation to identify the DEGs between RA and OA synovial tissues. Compared with previous studies that included a single dataset, our study adopted multiple datasets to get accurate and reliable results. We identified three hub genes, CXCL10, CXCL13, and ADIPOQ, which were validated using qRT-PCR. These genes may serve as novel biomarkers and potential targets for the difference of accurate diagnosis and treatment strategies between RA and OA. Further experiments are needed to verify these candidate biomarkers.
References
McInnes IB (2011) The pathogenesis of rheumatoid arthritis. N Engl J Med 365:2205–2219. https://doi.org/10.1056/NEJMra1004965
Scott DL, Wolfe F, Huizinga TW (2010) Rheumatoid arthritis. 376:1094–1108. https://doi.org/10.1016/S0140-6736(10)60826-4
Smolen JS, Aletaha D, Barton A et al (2018) Rheumatoid arthritis. Nat Rev Dis Primer 4:18001. https://doi.org/10.1038/nrdp.2018.1
Buckwalter J, Martin J (2006) Osteoarthritis. Adv Drug Deliv Rev 58:150–167. https://doi.org/10.1016/j.addr.2006.01.006
Felson DT (2000) Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med 133:635. https://doi.org/10.7326/0003-4819-133-8-200010170-00016
Lindblom A, Robinson PN (2011) Bioinformatics for human genetics: promises and challenges. Hum Mutat 32:495–500. https://doi.org/10.1002/humu.21468
Cai W, Li H, Zhang Y, Han G (2020) Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ 8:e8390. https://doi.org/10.7717/peerj.8390
Liang W, Sun F (2018) Identification of key genes of papillary thyroid cancer using integrated bioinformatics analysis. J Endocrinol Invest 41:1237–1245. https://doi.org/10.1007/s40618-018-0859-3
Liu L, Chen F, Xiu A et al (2018) Identification of key candidate genes and pathways in endometrial cancer by integrated bioinformatical analysis. Asian Pac J Cancer Prev 19. https://doi.org/10.22034/APJCP.2018.19.4.969
Han B, Feng D, Yu X et al (2018) Identification and interaction analysis of molecular markers in colorectal cancer by integrated bioinformatics analysis. Med Sci Monit 24:6059–6069. https://doi.org/10.12659/MSM.910106
Lauwerys BR, Hernández-Lobato D, Gramme P et al (2015) Heterogeneity of synovial molecular patterns in patients with arthritis. PLoS One 10:e0122104. https://doi.org/10.1371/journal.pone.0122104
Ungethuem U, Haeupl T, Witt H et al (2010) Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol Genomics 42A:267–282. https://doi.org/10.1152/physiolgenomics.00004.2010
Huber R, Hummert C, Gausmann U et al (2008) Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther 10:R98. https://doi.org/10.1186/ar2485
Woetzel D, Huber R, Kupfer P et al (2014) Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther 16:R84. https://doi.org/10.1186/ar4526
Ritchie ME, Phipson B, Wu D et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:13. https://doi.org/10.1093/nar/gkv007
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. https://doi.org/10.1038/75556
Kanehisa M, Furumichi M, Tanabe M et al (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1093/nar/gkw1092
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J Integr Biol 16:284–287. https://doi.org/10.1089/omi.2011.0118
Hadley W (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon P (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Demoruelle MK, Deane KD, Holers VM (2014) When and where does inflammation begin in rheumatoid arthritis? Curr Opin Rheumatol 26:64–71. https://doi.org/10.1097/BOR.0000000000000017
Kumar H, Bot A (2018) In this issue: role of immune cells and molecules in rheumatoid arthritis pathogenesis and cancer immunotherapy. Int Rev Immunol 37:127–128. https://doi.org/10.1080/08830185.2018.1469353
Chang WH, Lai AG (2019) The pan-cancer mutational landscape of the PPAR pathway reveals universal patterns of dysregulated metabolism and interactions with tumor immunity and hypoxia. Ann N Y Acad Sci 1448:65–82. https://doi.org/10.1111/nyas.14170
Hu X, Tang J, Zeng G et al (2019) RGS1 silencing inhibits the inflammatory response and angiogenesis in rheumatoid arthritis rats through the inactivation of Toll-like receptor signaling pathway. J Cell Physiol 234:20432–20442. https://doi.org/10.1002/jcp.28645
Kwak HB, Ha H, Kim H-N et al (2008) Reciprocal cross-talk between RANKL and interferon-γ–inducible protein 10 is responsible for bone-erosive experimental arthritis. Arthritis Rheum 58:1332–1342. https://doi.org/10.1002/art.23372
Antonelli A, Ferrari SM, Giuggioli D et al (2014) Chemokine (C–X–C motif) ligand (CXCL)10 in autoimmune diseases. Autoimmun Rev 13:272–280. https://doi.org/10.1016/j.autrev.2013.10.010
Bugatti S, Manzo A, Vitolo B et al (2014) High expression levels of the B cell chemoattractant CXCL13 in rheumatoid synovium are a marker of severe disease. Rheumatology 53:1886–1895. https://doi.org/10.1093/rheumatology/keu163
Shang H, Hao Y, Hu W et al (2019) Association between ADIPOQ gene variants and knee osteoarthritis in a Chinese population. Biosci Rep 39:BSR20182104. https://doi.org/10.1042/BSR20182104
Li Z, Zhong L, Du Z et al (2019) Network analyses of differentially expressed genes in osteoarthritis to identify hub genes. BioMed Res Int 2019:1–9. https://doi.org/10.1155/2019/8340573
Jiang L, Zhu X, Rong J et al (2018) Obesity, osteoarthritis and genetic risk: the rs182052 polymorphism in the ADIPOQ gene is potentially associated with risk of knee osteoarthritis. Bone Jt Res 7:494–500. https://doi.org/10.1302/2046-3758.77.BJR-2017-0274.R1
Fernández-Torres J, Martínez-Nava GA, Zamudio-Cuevas Y et al (2019) Epistasis between ADIPOQ rs1501299 and PON1 rs662 polymorphisms is potentially associated with the development of knee osteoarthritis. Mol Biol Rep 46:2049–2058. https://doi.org/10.1007/s11033-019-04654-5
Funding
This work was funded by the National Natural Science Funds of China (81774114, 82074234), Shanghai Chinese Medicine Development Office, Shanghai Chinese and Western Medicine Clinical Pilot Project (ZY(2018–2020)-FWTX-1010), Shanghai Chinese Medicine Development Office, Shanghai Traditional Chinese Medicine Specialty Alliance Project (ZY(2018–2020)-FWTX-4017), and National Administration of Traditional Chinese Medicine, Regional Chinese Medicine (Specialist) Diagnosis and Treatment Center Construction Project-Rheumatology.
Author information
Authors and Affiliations
Contributions
Dongyi He conceived the content. Runrun Zhang and Xinpeng Zhou drafted the manuscript. Yehua Jin and Cen Chang contributed to statistical data analysis and qPCR verification. Rongsheng Wang, Junyu Fan, and Jia Liu checked the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of Guanghua Hospital of Integrated Traditional Chinese and Western Medicine (approval number: 2018-K-12) and written consent was collected before the surgery from the patients.
Consent to participate
The authors consented to their participation in the research.
Consent for publication
All authors read and approved the final manuscript.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, R., Zhou, X., Jin, Y. et al. Identification of differential key biomarkers in the synovial tissue between rheumatoid arthritis and osteoarthritis using bioinformatics analysis. Clin Rheumatol 40, 5103–5110 (2021). https://doi.org/10.1007/s10067-021-05825-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05825-1