Abstract
Vitamin D is involved in immune system modulation as well as in calcium and bone homeostasis, hence plays a role in rheumatoid arthritis (RA) etiopathogenesis. A bulk of studies in different populations have assessed the association between the vitamin D receptor (VDR) gene polymorphisms and the risk of RA, reporting conflicting results. Therefore, we designed a meta-analysis to comprehensively evaluate the association of VDR gene polymorphisms and RA risk. All potential studies reporting the association between VDR gene polymorphisms and susceptibility to RA published till February 2020 were retrieved through systematic search of database, including Scopus and MEDLINE. Strength of pooled association was determined through calculating the pooled odds ratios (ORs) and 95% confidence intervals (CIs). Subgroup analysis was performed by stratifying the studies by population type. This meta-analysis included 23 eligible studies (21 articles) overall. We noticed that FokI SNP had a significant protective association with susceptibility to RA in the overall analysis as well as in Europeans and Asians. TaqI SNP decreased the RA risk in Africans and Arabs, but not in the overall analysis. Likewise, BsmI SNP and RA risk in the overall population analysis was not significant. Interestingly, BsmI polymorphism increased RA risk in Africans. This meta-analysis offers a significant association between VDR gene polymorphism and susceptibility to RA in both overall and ethnic-specific analysis. However, different polymorphisms acted inversely in increasing or decreasing RA risk in different populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Rheumatoid arthritis (RA) is a chronic inflammatory disorder, which is characterized by production of autoantibody, chronic synovial inflammation, and progressive destruction and deformity of joint [1,2,3]. RA is a devastating and common autoimmune disease that has a prevalence of approximately 0.3% to 1% of the total population and more frequently occurs in women than in men (3:1 ratio) [4, 5]. While the main etiology of RA is yet unknown, several population base studies have reported that genetic susceptibility and environmental factors play a principal role in the onset and progression of the disorder [6]. Early investigations proposed that genetic factors contribute to about 50–65% of the RA developing risk [7]. Human leukocyte antigen (HLA) is one of the common significant genetic loci for RA susceptibility [8, 9]. However, family studies recommend that the HLA region is attributed to only about 30% of genetic susceptibility and that non-HLA loci, such as Cytotoxic T lymphocyte-associated protein 4 (CTLA4), Peptidyl arginine deiminase 4 (PADI4), Methylenetetrahydrofolate reductase (MTHFR), TIM (T cell/transmembrane, immunoglobulin, and mucin) gene family, and Tumor necrosis factor alpha-induced protein 3 (TNFAIP3), have also been associated with RA predisposition [10,11,12,13,14,15,16].
Vitamin D is a steroid-like hormone that acts by binding to vitamin D receptor (VDR), belonging to the nuclear hormone receptor superfamily. Vitamin D/VDR signaling plays an important role in the regulation of immune cell proliferation and differentiations, lymphocyte activation, and cytokine production, probably contributing to autoimmunity [17, 18]. Other than its critical function in the calcium metabolism and bone homeostasis, vitamin D plays an immunomodulatory role [19]. It also exhibits anti-inflammatory and anti-infection characteristics [20, 21]. Several studies have suggested that VDR signaling plays a critical function in T-cell differentiation and function. Widespread investigations have demonstrated the involvement of T cells in the etiopathogenesis of RA [22]. Genetic variation of the VDR gene and abnormal levels of vitamin D could result in the initiation and perpetuation of multiple autoinflammatory disorders like RA [23, 24].
It has been demonstrated that the biological function of vitamin D can be affected by single-nucleotide polymorphisms (SNPs) of the VDR gene [25]. Recently, VDR gene SNPs have become the focus of association studies in searching for genetic factors involved in the RA risk. On the other side, although the functional significances of VDR polymorphisms remain obscure, these VDR gene polymorphisms have been associated with an increased risk of several autoimmune and inflammatory diseases, such as type 1 diabetes (T1D), multiple sclerosis (MS), and asthma [26,27,28]. The four commonly studied VDR polymorphisms sites are BsmI, ApaI, FokI, and TaqI. BsmI and ApaI polymorphisms are located near the 3′ end of the VDR gene in the intron between exons 8 and 9, and TaqI is located in exon 9. These polymorphisms result in silent codon mutations associated with raised VDR mRNA stability [29]. FokI polymorphism is located in exon 2 and leads to the production of a protein with different sizes; the smaller form of the protein (424 amino acids) is more effective than the long form (427 amino acids) [30].
Association studies between VDR gene polymorphisms and risk of RA conducted in multiple populations have yielded conflicting results; some revealed significant correlation while other studies failed to reach statistical significance [31,32,33]. The causes for this discrepancy may be due to low statistical power, sample sizes and/or clinical heterogeneity. To offset these limitations, we conducted this most up-to-date meta-analysis to evaluate whether VDR gene polymorphisms are associated with RA susceptibility.
Methods
The current meta-analysis follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [34]. No ethics committee approval was necessary for this meta-analysis, which does not contain any studies with human participants or animals performed by any of the authors.
Literature search
A comprehensive systematic search was conducted in Scopus and MEDLINE databases and retrieved all relevant publications till February 2020 (the search was also updated before submission). The applied key words for search were as follows: (“Rheumatoid Arthritis” OR “Arthritis” OR “RA”) AND (“VDR” OR “vitamin D receptor”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). The reference list of all studies was cross-checked to find other potential studies which might have been missed during initial search.
Study selection criteria
Our initial search strategy yields 233 studies that were exported to EndNote X8. The title and abstract of all studies were scanned by two authors and irrelevant studies were excluded. Full-text verification was performed if we could not categorize studies based on their title and abstract. Any disagreements during study selection was discussed and resolved by consensus.
Inclusion and exclusion criteria
The primary search results were transported to the EndNote and publications were screened based on the following criteria: (1) all publications considering the association between VDR gene polymorphisms (FokI (rs2228570) or/and TaqI (rs731236) or/and BsmI (rs1544410) or/and ApaI (rs7975232)) and RA risk; (2) all observational studies (cohort or case–control design); (3) publications with sufficient data to extract or calculate odds ratios (ORs) and 95% confidence intervals (CIs); (4) publications that report genotype or allele frequencies in RA patients and healthy individuals. Reviews, meta-analysis, case reports, book chapters, letters to the editor, conference abstracts, as well as duplicates were all excluded. The application of these criteria recognized 15, 11, 17, and 9 eligible studies for FokI, TaqI, BsmI, and ApaI polymorphisms, respectively.
Data extraction and quality assessment
All required data were extracted conforming to the standardized extraction checklist for the following data: the first author’s name, journal and year of publication, country of origin, ethnicity, number of subjects in the case and control groups, mean or range of age, genotyping method, and genotype counts in the case and control group. In order to improve the accuracy of our data, two authors independently extract data and possible discrepancies were solved by consensus. In the current meta-analysis, we exploited the Newcastle–Ottawa Scale (NOS) to assess methodological quality of included studies [35]. Collectively, we divided publications into three groups: higher quality score ≥ 7; moderate quality 4 ≤ score < 7; low quality score < 4.
Statistical analysis
Deviation from Hardy–Weinberg equilibrium (HWE) for distribution of the allele frequencies was analyzed by χ2-test in control groups. The association between VDR gene polymorphism and RA was assessed by estimating ORs and their corresponding 95% CIs. For each SNP, the dominant model, recessive model, allelic model, homozygous model, and heterozygous model were examined to estimate its effect. In detail, defined models for FokI, TaqI, BsmI, and ApaI SNPs are as follows: FokI—dominant model (ff + Ff vs. FF), recessive model (ff vs. Ff + FF), allelic model (f vs. F), homozygote (ff vs. FF), and heterozygote (Ff vs. FF); TaqI—dominant model (tt + Tt vs. TT), recessive model (tt vs. Tt + TT), allelic model (t vs. T), homozygote (tt vs. TT), and heterozygote (Tt vs. TT); BsmI—dominant model (bb + Bb vs. BB), recessive model (bb vs. Bb + BB), allelic model (b vs. B), homozygote (bb vs. BB), and heterozygote (Bb vs. BB); ApaI—dominant model (aa + Aa vs. AA), recessive model (aa vs. Aa + AA), allelic model (a vs. A), homozygote (aa vs. AA), and heterozygote (Aa vs. AA). The heterogeneity among studies was measured by the χ2 test–based Q and I2 value which quantify the degree of heterogeneity [36]. In the case of heterogeneity (Q statistic with a P value less than 0.1 and I2 exceeding 50%), random-effects model (REM) was employed [37]; otherwise, fixed-effect model was exploited [38]. Potential publication bias was estimated by Egger’s linear regression test, and Begg’s test (P value < 0.05 considered statistically significant) [39]. Finally, we utilized sensitivity analysis to show the stability of our results. All statistical tests for this meta-analysis were performed with Stata statistical software (version 14.0; Stata Corporation, College Station, TX, USA) and SPSS (version 23.0; SPSS, Inc. Chicago, IL, USA).
Results
Study characteristics
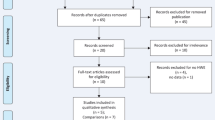
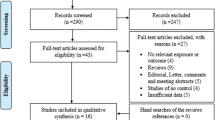
The four-phase process of study selection based on the PRISMA statement is outlined in Fig. 1. After the removal of duplicates (50 publications), 183 publications remained. Of these, 124 publications were excluded based on the title and abstract screening, and 36 publications were excluded by full-text evaluation. Ultimately, 23 eligible studies (21 articles) were included in final analysis [31,32,33, 40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57]. The references of all eligible publications were cross-checked and no more study was found. The studies were published between 2001 and 2019 and had an overall good methodological quality with NOS scores ranging from 6 to 8. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and TaqMan were used by majority of the included studies as genotyping method. Tables 1 and 2 summarize the characteristics and genotype frequency of the included studies.
Quantitative synthesis
In the current meta-analysis, FF for FokI SNP, TT for TaqI SNP, BB for BsmI SNP, and AA for ApaI SNP were used as the reference category.
Meta-analysis of FokI (rs2228570) SNP and RA risk
For FokI SNP, 15 case–control studies (13 articles) with 2170 cases and 2452 controls were included in quantitative analysis [31,32,33, 42, 44, 46, 47, 49, 52,53,54, 56, 57]. Of them, seven studies were performed in Europe, four studies were in Asia, three studies were in Africa, and only one study was carried out in the USA. The pooled OR of overall population detected a significant protective association between FokI SNP and susceptibility to RA under the dominant model (OR = 0.74, 95% CI = 0.60–0.92, P < 0.001), the ff versus FF model (OR = 0.66, 95% CI = 0.54–0.81, P < 0.001), and the Ff versus FF model (OR = 0.85, 95% CI = 0.73–0.98, P < 0.001), but not the allelic model (OR = 0.96, 95% CI = 0.42–1.14, P = 0.14) and the recessive model (OR = 0.54, 95% CI = 0.29–1.04, P = 0.06) (Fig. 2). For more clarifications, subgroup analysis by ethnicity was performed. The analyses showed a remarkable decreased risk of RA in Europeans across all genotype models (Fig. 3). In addition, Asians showed a decreased risk of RA under the dominant (OR = 0.65, 95% CI = 0.48–0.89, P < 0.001), Ff versus FF (OR = 0.56, 95% CI = 0.35–0.89, P = 0.01), and Ff versus FF (OR = 0.69, 95% CI = 0.50–0.96, P = 0.02) models, but not the recessive and allelic models. No significant associations were found in Africans and Arabs (Table 3).
Meta-analysis of TaqI (rs731236) SNP and RA risk
There were 11 case–control studies containing 1334 cases and 1560 controls concerning TaqI polymorphism and RA risk [31, 32, 40,41,42, 50, 52,53,54,55,56]. Of those, four studies were conducted in Europeans, four studies were in Asians, and three studies were in Africans. There was no evidence of significant association between TaqI polymorphism and RA risk in the pooled results. However, subgroup analyses indicated interesting results. In this regard, our analysis revealed a protective role of TaqI polymorphism in Africans and Arabs. In Africans, all of the genetic models, including the dominant (OR = 0.50, 95% CI = 0.29–0.85, P = 0.01), recessive (OR = 0.44, 95% CI = 0.25–0.79, P < 0.001), allelic (OR = 0.57, 95% CI = 0.37–0.88, P = 0.01), tt versus TT (OR = 0.32, 95% CI = 0.15–0.72, P < 0.001), and Tt versus TT (OR = 0.57, 95% CI = 0.38–0.87, P < 0.001) models were associated with decreased risk of RA. Furthermore, statistically significant and protective association of the recessive (OR = 0.53, 95% CI = 0.32–0.87, P = 0.01) and tt versus TT (OR = 0.43, 95% CI = 0.20–0.94, P = 0.03) models were detected in Arabs. No significant association was detected for Europeans and Asians (Table 3).
Meta-analysis of BsmI (rs1544410) SNP and RA risk
For BsmI SNP, 17 case–control studies (16 articles) encapsulating 2153 cases and 2326 controls subjects examined the association between BsmI polymorphism and RA risk [31,32,33, 40,41,42,43, 45,46,47,48, 51,52,53,54, 56]. Among included studies, seven studies were conducted in Europeans, six studies were in Asians, and four studies were in Africans. Our findings did not indicate any association between BsmI SNP and RA risk in the overall population analysis. Nonetheless, subgroup analysis found a significant positive association between BsmI SNP and RA risk in Africans under all genetic models: the dominant model (OR = 1.82, 95% CI = 1.14–2.88 2, P = 0.01), the recessive model (OR = 1.77, 95% CI = 1.13–2.78, P = 0.01), the allelic model (OR = 1.59, 95% CI = 1.14–2.23, P < 0.001), the bb versus BB model (OR = 2.40, 95% CI = 1.22–4.71, P = 0.01), and the Bb versus BB model (OR = 1.45, 95% CI = 1.04–2.01, P = 0.02) (Fig. 3). No significant association was detected for Europeans, Asians, and Arabs (Table 3).
Meta-analysis of ApaI (rs7975232) SNP and RA risk
Herein, nine studies were found providing data about ApaI polymorphism and RA risk [32, 40, 47, 48, 50, 52,53,54, 56]. A total of 1191 cases and 1415 controls were included in the quantitative analysis. Of eligible studies, four studies were performed in Asians, three studies were in Africans, and two studies were in Europeans. The analyses revealed no association between ApaI SNP and RA risk across all models in both overall population and subgroups. However, this polymorphism was significantly associated with RA risk under the Aa versus AA model in the overall analysis (OR = 0.76, 95% CI = 0.61–0.94, P = 0.01) (Table 3).
Evaluation of heterogeneity and publication bias
The degree of heterogeneity was measured for all five genetic models among intended genes. Collectively, significant heterogeneity was observed for FokI, TaqI, BsmI, and ApaI genes and subsequently random-effect model was used (Table 3). The Egger regression, Begg rank correlation analysis, and funnel plot according to quantitative analysis demonstrated no statistical significance (Table 3, Fig. 4).
Sensitivity analysis
Sensitivity analysis was performed by removing a single study at a time to evaluate the robustness of the results. Accordingly, the significance of the pooled ORs was not affected by any single study in the dominant model for FokI, TaqI, BsmI, and ApaI SNPs (Fig. 5).
Discussion
In the current most up-to-date systematic review and meta-analysis study, we intended to obtain a conclusive and exact estimation of the associations between the polymorphisms located on the VDR gene, including FokI (rs2228570), BsmI (rs1544410), TaqI (rs731236), and ApaI (rs7975232) and risk of RA predisposition. The results of the meta-analysis on 23 eligible studies (21 articles) unraveled that FokI and TaqI polymorphisms in the overall and subgroup analysis, respectively, had significant association with RA risk.
During the past years, numerous investigations have evaluated the association of VDR gene polymorphisms and risk of RA throughout different populations. That notwithstanding, these studies sometimes confirmed findings in different populations, but sometimes not. The conflict among these studies may be due to differences in the genotyping methods, clinical heterogeneity of the patients, variations in the diagnosis of patients, small sample sizes, lack of statistical power, and the interactions between genetic content and environmental risk factors offered by different geographic regions. As a result, three previous meta-analyses have tried to settle the issue [58,59,60]. That notwithstanding, several original association studies investigated the association of VDR gene polymorphisms and RA risk, after the latest meta-analysis published in 2016. As a consequence, it seems paramount to perform an up-to-date meta-analysis to achieve more valid and comprehensive pooled approximation on the association of VDR gene SNPs and RA risk.
The meta-analysis was performed by Tizaoui et al. in 2015 [59]; 12 case–control studies, involving 1703 cases and 2635 controls, were included. This analysis indicated a significant association between TaqI polymorphism and RA disease in homozygous, codominant, and allele contrast models. Association between BsmI polymorphism and RA risk was marginal in the dominant, codominant, and allele contrast models. The association between FokI polymorphism and RA risk was significant in the recessive, dominant, and allele contrast models. Subgroup analysis indicated that publication year, ethnicity, age, latitude, and estimated 25(OH)D levels influenced significantly the association between VDR polymorphisms and RA risk. Therefore, study characteristics impressed the association between VDR gene polymorphisms and RA disease. This meta-analysis suggested that VDR gene TaqI and FokI polymorphisms are involved in RA risk. On the other side, in the last meta-analysis conducted by Song et al. in 2016, seven studies containing a total of 923 patients and 912 controls were included in the meta-analysis, and three of the VDR gene SNPs, FokI, BsmI, and TaqI, were considered [60]. This study found no association between FokI, BsmI, and TaqI polymorphisms and risk of RA in the overall analysis. However, FokI SNP was associated with increased risk of RA in Europeans (OR = 1.40). Our most up-to-date meta-analysis included 23 eligible studies (21 articles) in total, 15 case–control studies (13 articles) with 2170 cases and 2452 controls for FokI SNP, 11 case–control studies containing 1334 cases and 1560 controls for TaqI polymorphism, 17 case–control studies (16 articles) involving 2153 cases and 2326 controls for BsmI SNP, and 9 studies containing 1191 cases and 1415 controls for ApaI polymorphism. We indicated that FokI SNP had a significant association with susceptibility to RA and was protective under the dominant model (OR = 0.74), the ff versus FF model (OR = 0.66), and the Ff versus FF model (OR = 0.85). Moreover, this polymorphism decreased the RA risk in Europeans in all models and also was protective in Asians under the dominant, Ff versus FF, and Ff versus FF models. On the other hand, although we did not detect a significant association of TaqI SNP and RA risk in the overall analysis, this polymorphism decreased the RA risk in Africans and Arabs. As such, BsmI SNP and RA risk in the overall population analysis was not significant. Nonetheless, this polymorphism increased RA risk in Africans under all genetic models. Finally, neither overall nor subgroup analyses indicated association of ApaI SNP with RA risk. The differences in the findings of the current meta-analysis with the previous ones may stem from the difference in the sample size and ethnicity, as our meta-analysis included further studies with diverse populations.
The interaction of common polymorphisms might be involved in determining the genetic etiopathogenesis of the multifactorial diseases. SNPs have been reported to cause little, but rarely significant, biological impact on the protein they are encoding [61]. VDR gene polymorphisms, such as ApaI, BsmI, and TaqI, have been suggested to lack considerable influence on the protein structure of VDR. Nevertheless, these SNPs may modulate the VDR protein stability, translation quality, or splicing of the corresponding mRNA. Interestingly, FokI SNP has been reported to modify the VDR protein structure and the efficacy of mRNA transcription [62]. Hence, VDR gene ApaI, BsmI, and TaqI SNPs may be associated with diseases pathogenesis during the linkage disequilibrium with the real disease-associating genes [63]. Environmental interactions and ethnicity of the population may be critical in determining the function and expression of VDR [64], which are regarded as confounding factors during the association studies.
It should be noted that our meta-analysis was not bereft of limitations and caveats. First, we searched only English-written papers, which may raise the possibility of omission of potentially valuable studies. Second, we did not analyze the role and effect of age, lifestyle, gender, and other genetic SNPs as confounding factors on the association of VDR gene SNPs and RA risk. As a consequence, more investigations about the gene–environment and gene–gene interactions are still indispensable to attain an exhausted estimation of VDR gene polymorphism with RA risk. Third, we observed a significant heterogeneity among the studies for different genetic models in the SNPs, which may affect the way that the findings are interpreted. Finally, there are other polymorphisms in the VDR gene that have been studied in RA patients, but could not be involved in this meta-analysis because of insufficient amount of data. As a result, the results should be interpreted with caution.
Conclusion
All in all, this was the most up-to-date meta-analysis currently with respect to the association of VDR gene SNPs with RA risk. We evaluated 23 eligible studies (21 articles) to uncover the bona fide association of VDR gene FokI (rs2228570), TaqI (rs731236), BsmI (rs1544410), and ApaI (rs7975232) polymorphisms with risk of RA susceptibility. We indicated that FokI SNP had a significant protective association with susceptibility to RA in the overall analysis as well as in Europeans and Asians. TaqI SNP decreased the RA risk in Africans and Arabs, but not in the overall analysis. As such, BsmI SNP and RA risk in the overall population analysis was not significant. Interestingly, BsmI polymorphism increased RA risk in Africans. Further studies on the VDR gene in RA patients other than the genetic as well as traditional risk factors may provide a possibility for recognizing the important susceptibility factors in the RA development, which might be used in the personalized medicine for better and optimized therapy of RA patients.
References
McInnes IB, Schett G (2011) The pathogenesis of rheumatoid arthritis. N Engl J Med 365(23):2205–2219
Abbasi M, Mousavi MJ, Jamalzehi S, Alimohammadi R, Bezvan MH, Mohammadi H, Aslani S (2019) Strategies toward rheumatoid arthritis therapy; the old and the new. J Cell Physiol 234(7):10018–10031
Mousavi MJ, Jamshidi A, Chopra A, Aslani S, Akhlaghi M, Mahmoudi M (2019) Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J Cell Physiol 234(1):335–347
Sangha O (2000) Epidemiology of rheumatic diseases. Rheumatology 39(suppl_2):3–12
Tobón GJ, Youinou P, Saraux A (2010) The environment, geo-epidemiology, and autoimmune disease: rheumatoid arthritis. Autoimmun Rev 9(5):A288–A292
Conigliaro P, Triggianese P, de Martino E, Fonti GL, Chimenti MS, Sunzini F, Viola A, Canofari C, Perricone R (2019) Challenges in the treatment of rheumatoid arthritis. Autoimmun Rev 18:706–713
Orozco G, Barton A (2010) Update on the genetic risk factors for rheumatoid arthritis. Expert Rev Clin Immunol 6(1):61–75
Holoshitz J (2010) The rheumatoid arthritis HLA-DRB1 shared epitope. Curr Opin Rheumatol 22(3):293–298
Karami J, Aslani S, Jamshidi A, Garshasbi M, Mahmoudi M (2019) Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 702:8–16
Gonzalez-Escribano M et al (1999) CTLA4 polymorphisms in Spanish patients with rheumatoid arthritis. Tissue Antigens 53(3):296–300
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG (2007) PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int 27(9):827–833
Berkun Y, Levartovsky D, Rubinow A, Orbach H, Aamar S, Grenader T, Abou Atta I, Mevorach D, Friedman G, Ben-Yehuda A (2004) Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann Rheum Dis 63(10):1227–1231
Razi B et al (2019) TIM family gene polymorphism and susceptibility to rheumatoid arthritis: systematic review and meta-analysis. PLoS One 14(2):e0211146
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2012) Associations between TNFAIP3 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm Res 61(6):635–641
Ebrahimiyan H, Mostafaei S, Aslani S, Jamshidi A, Mahmoudi M (2019) Studying the association between STAT4 gene polymorphism and susceptibility to rheumatoid arthritis disease: an updated meta-analysis. Iranian J Immunol 16(1):71–83
Mohammadi SA et al (2018) IL27 gene single nucleotide polymorphisms confer susceptibility to rheumatoid arthritis in Iranian population. Meta Gene 18:149–152
Kalueff A et al (2006) The vitamin D neuroendocrine system as a target for novel neurotropic drugs. CNS Neurol Disord Drug Targets 5(3):363–371
Adorini L (2002) Immunomodulatory effects of vitamin D receptor ligands in autoimmune diseases. Int Immunopharmacol 2(7):1017–1028
Ranganathan P (2009) Genetics of bone loss in rheumatoid arthritis—role of vitamin D receptor polymorphisms. Rheumatology 48(4):342–346
Sun J (2010) Vitamin D and mucosal immune function. Curr Opin Gastroenterol 26(6):591–595
Tian Y, Wang C, Ye Z, Xiao X, Kijlstra A, Yang P (2012) Effect of 1, 25-dihydroxyvitamin D3 on Th17 and Th1 response in patients with Behcet's disease. Invest Ophthalmol Vis Sci 53(10):6434–6441
Nagpal S, Na S, Rathnachalam R (2005) Noncalcemic actions of vitamin D receptor ligands. Endocr Rev 26(5):662–687
Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A (2006) Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. Jama 296(23):2832–2838
Shoenfeld N, Amital H, Shoenfeld Y (2009) The effect of melanism and vitamin D synthesis on the incidence of autoimmune disease. Nat Clin Pract Rheumatol 5(2):99–105
Uitterlinden AG et al (2004) Vitamin D receptor gene polymorphisms in relation to vitamin D related disease states. J Steroid Biochem Mol Biol 89:187–193
Makoui, M.H., Vitamin D receptor gene polymorphisms and the risk of the type 1 diabetes: a meta-analysis based on 40 case–control studies. Department of Immunology, School of Public Health, Tehran University of …
Imani D, Razi B, Motallebnezhad M, Rezaei R (2019) Association between vitamin D receptor (VDR) polymorphisms and the risk of multiple sclerosis (MS): an updated meta-analysis. BMC Neurol 19(1):339
Makoui MH, Imani D, Motallebnezhad M, Azimi M, Razi B (2020) Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case–control studies. Ann Allergy Asthma Immunol 124(1):57–69
Uitterlinden AG, Fang Y, van Meurs JBJ, Pols HAP, van Leeuwen JPTM (2004) Genetics and biology of vitamin D receptor polymorphisms. Gene 338(2):143–156
Sundqvist E, Bäärnhielm M, Alfredsson L, Hillert J, Olsson T, Kockum I (2010) Confirmation of association between multiple sclerosis and CYP27B1. Eur J Hum Genet 18(12):1349–1352
Maalej A, Petit-Teixeira E, Michou L, Rebai A, Cornelis F, Ayadi H (2005) Association study of VDR gene with rheumatoid arthritis in the French population. Genes Immun 6(8):707–711
Mosaad YM, Hammad EM, Fawzy Z, Abdal Aal IA, Youssef HM, ElSaid TO, Monir R, el-Deek BS (2014) Vitamin D receptor gene polymorphism as possible risk factor in rheumatoid arthritis and rheumatoid related osteoporosis. Hum Immunol 75(5):452–461
Ghelani AM, Samanta A, Jones AC, Mastana SS (2011) Association analysis of TNFR2, VDR, A2M, GSTT1, GSTM1, and ACE genes with rheumatoid arthritis in south Asians and Caucasians of East Midlands in the United Kingdom. Rheumatol Int 31(10):1355–1361
Moher D, Liberati A, Tetzlaff J, Altman DG (2010) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg 8(5):336–341
Stang A (2010) Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9):603–605
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088–1101
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Bmj 315(7109):629–634
Garcia-Lozano J et al (2001) Association of vitamin D receptor genotypes with early onset rheumatoid arthritis. Eur J Immunogenet 28(1):89–93
Lee C-K, Hong JS, Cho YS, Yoo B, Kim GS, Moon HB (2001) Lack of relationship between vitamin D receptor polymorphism. J Korean Med Sci 16:188–192
Goertz B, Fassbender WJ, Williams JC, Marzeion AM, Bretzel RG, Stracke H, Berliner MN (2003) Vitamin D receptor genotypes are not associated with rheumatoid arthritis or biochemical parameters of bone turnover in German RA patients. Clin Exp Rheumatol 21(3):333–339
Rass P, Pákozdi A, Lakatos P, Zilahi E, Sipka S, Szegedi G, Szekanecz Z (2006) Vitamin D receptor gene polymorphism in rheumatoid arthritis and associated osteoporosis. Rheumatol Int 26(11):964–971
Hitchon CA et al (2012) Vitamin D receptor polymorphism rs2228570 (Fok1) is associated with rheumatoid arthritis in North American natives. J Rheumatol 39(9):1792–1797
Hussien YM, Shehata A, Karam RA, Alzahrani SS, Magdy H, el-Shafey AM (2013) Polymorphism in vitamin D receptor and osteoprotegerin genes in Egyptian rheumatoid arthritis patients with and without osteoporosis. Mol Biol Rep 40(5):3675–3680
Karray EF, Ben Dhifallah I, Ben Abdelghani K, Ben Ghorbel I, Khanfir M, Houman H, Hamzaoui K, Zakraoui L (2012) Associations of vitamin D receptor gene polymorphisms FokI and BsmI with susceptibility to rheumatoid arthritis and Behçet's disease in Tunisians. Joint Bone Spine 79(2):144–148
Y, H., Associations of vitamin D receptor gene polymorphisms with RA in the Han population of human in China. Zhong Nan Da Xue Xue, 2013: p. 310–316
Li C, W.Y., Yin H and Yin S, The association between vitamin D, vitamin D receptor polymorphism and rheumatoid arthritis. Chinese Journal of Rheumatology 2013(17): p. 164–168
Shukla S et al (2014) Role of PTPN22 and VDR gene polymorphisms in susceptibility to rheumatoid arthritis: a study from Central India. Adv Genom Genet 4:79–85
Tizaoui K, Kaabachi W, Ouled Salah M, Ben Amor A, Hamzaoui A, Hamzaoui K (2014) Vitamin D receptor TaqI and ApaI polymorphisms: a comparative study in patients with Behçet’s disease and rheumatoid arthritis in Tunisian population. Cell Immunol 290(1):66–71
John P, Bhatti A, Ain N, Iqbal T, Sadaf T, Malik JM (2017) Case–control study of vitamin D receptor gene polymorphism in Pakistani rheumatoid arthritis patients. Rev Bras Reumatol 57(6):633–636
Saad, M.N., et al., Genetic case-control study for eight polymorphisms associated with rheumatoid arthritis. PLoS One, 2015. 10(\)
Di Spigna G et al (2016) Vitamin D receptor polymorphisms as tool for early screening of severe bone loss in women patients with rheumatoid arthritis. Eur Rev Med Pharmacol Sci 20(22):4664–4669
Khoja SO, el-Miedany Y, Iyer AP, Bahlas SM, Balamash KS, Elshal MF (2018) Associations of vitamin D levels and vitamin D receptor genotypes with patient-reported outcome/disease activity in patients with rheumatoid arthritis. Clin Lab 64(1):51–58
Mahmoud SAAH et al (2018) Vitamin D deficiency and RS731236 (taq1) vitamin D receptor gene polymorphism as possible risk factors for rheumatoid arthritis and osteoarthritis. Acta Med Austriaca 34:209
Mukhtar M, Sheikh N, Suqaina SK, Batool A, Fatima N, Mehmood R, Nazir S (2019) Vitamin D receptor gene polymorphism: an important predictor of arthritis development. Biomed Res Int 2019:1–8
Rodríguez-Carrio J et al (2019) Vitamin D receptor polymorphism and DHCR7 contribute to the abnormal interplay between vitamin D and lipid profile in rheumatoid arthritis. Sci Rep 9(1):1–11
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2011) Associations between vitamin D receptor polymorphisms and susceptibility to rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep 38(6):3643–3651
Tizaoui K, Hamzaoui K (2015) Association between VDR polymorphisms and rheumatoid arthritis disease: systematic review and updated meta-analysis of case–control studies. Immunobiology 220(6):807–816
Song G, Bae S-C, Lee Y (2016) Vitamin D receptor FokI, BsmI, and TaqI polymorphisms and susceptibility to rheumatoid arthritis. Z Rheumatol 75(3):322–329
Vinh Quốc Lương K, Nguyễn LTH (2012) The role of vitamin D in asthma. Pulm Pharmacol Ther 25(2):137–143
van Etten E, Verlinden L, Giulietti A, Ramos-Lopez E, Branisteanu DD, Ferreira GB, Overbergh L, Verstuyf A, Bouillon R, Roep BO, Badenhoop K, Mathieu C (2007) The vitamin D receptor gene FokI polymorphism: functional impact on the immune system. Eur J Immunol 37(2):395–405
Lemos MC, Fagulha A, Coutinho E, Gomes L, Bastos M, Barros L, Carrilho F, Geraldes E, Regateiro FJ, Carvalheiro M (2008) Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Hum Immunol 69(2):134–138
Vanessa O et al (2013) Vitamin D receptor gene expression and function in a South African population: ethnicity, vitamin D and FokI. PLoS One 8(6):e67663
Acknowledgments
We thank Dr. Bahman Razi for comments that greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
Z.B. generated the idea. D.I. analyzed and interpreted the data. Z.B. and H.Y. prepared the original draft. Z.B., H.Y., and M.A. critically revised the paper. M.A. supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bagheri-Hosseinabadi, Z., Imani, D., Yousefi, H. et al. Vitamin D receptor (VDR) gene polymorphism and risk of rheumatoid arthritis (RA): systematic review and meta-analysis. Clin Rheumatol 39, 3555–3569 (2020). https://doi.org/10.1007/s10067-020-05143-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-020-05143-y