Abstract
Mutations in the HINT1 gene were recently discovered as being the major cause of autosomal recessive axonal neuropathy with neuromyotonia. This combination was clinically recognized and described previously in a few reports but is generally unknown. We aimed to establish the importance of HINT1 mutations as the cause of hereditary neuropathy and particularly hereditary motor neuropathy/axonal Charcot-Marie-Tooth (HMN/CMT2) among Czech patients. Overall, mutations in the HINT1 gene seem to be a surprisingly frequent cause of inherited neuropathy in our group of patients. Biallelic pathogenic mutations were found in 21 patients from 19 families. The prevalent mutation in the Czech population is the p.R37P (95 % of pathogenic alleles). Clinically, all patients with biallelic mutations presented with early onset of symptoms at the end of the first decade. Foot/toe extension weakness to plegia was present in almost all patients. Neuromyotonia was present in all but two patients. However, it had been properly recognized in only three patients prior to molecular genetic diagnosis. HINT1 mutations seem to be one of the most frequent causes of inherited neuropathy and are probably the most frequent cause of HMN in Czech patients. We suggest all HMN/CMT2 patients be tested for the presence of the prevalent mutation, the p.R37P.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Charcot-Marie-Tooth (also called hereditary motor and sensory neuropathy (HMSN)) disease encompasses a heterogeneous group of disorders where diffuse polyneuropathy lies at the core of the problem. Clinically, it presents itself as muscle weakness, wasting, and sensory loss, usually most severe distally [1]. Genetically, mutations in more than 60 genes have been associated with Charcot-Marie-Tooth (CMT) to date. In some cases, mutations in specific genes might have specific clinical presentments.
Classification of hereditary neuropathies has undergone long development. Basic distinction relating to nerve pathophysiology is still very helpful however. Demyelinating inherited peripheral motor and sensory neuropathies are called CMT1; axonal motor and sensory inherited neuropathies are known as CMT2. CMT patients are divided into these two groups according to motor nerve conduction velocities (of the median or ulnar nerve), with the cutoff value being 38 m/s. Later, a group of intermediate inherited peripheral neuropathies was also identified. This group is a subgroup of disorders with nerve conduction velocities in between the demyelinating and the axonal groups.
Hereditary motor neuropathy (HMN) is a rarer form (about 10 %) of inherited peripheral neuropathies [2]. This group is characterized by pure motor axonal involvement and no sensory deficit and is also very heterogeneous, both clinically and genetically.
The axonal CMT (CMT2) group and the distal hereditary motor neuropathy (HMN) group clinically overlap, often dependent on the stage of the disease and the age of the patient. Based on current knowledge, it is possible to identify the molecular genetic cause of HMN in approx. 20 % of patients [3]. Therefore, a large portion of patients with HMN/CMT2 remain without proper molecular genetic diagnosis. Efforts to identify the cause of HMN/CMT2 disease are hence desirable.
Recently, mutations in the gene HINT1 have been shown to be associated with a specific CMT2/HMN disease entity: Charcot-Marie-Tooth with neuromyotonia also known as autosomal recessive peripheral neuropathy with neuromyotonia (ARAN-NM) [2, 4, 5]. M. Zimon et al. clarified the molecular cause of ARAN-NM [5]. In their report on a large cohort of ARAN-NM patients from different countries, mutations in the HINT1 gene were detected in 68 % of the cohort. The prevalent mutation has been recognized as p.R37P.
Neuromyotonia has been previously associated with axonal Charcot-Marie-Tooth or HMN [6–8]. It is defined as delayed muscular relaxation after a voluntary contraction resulting from a disorder of the peripheral nerve rather than the muscle [6]. The spontaneous engagement of peripheral motor nerves which gives rise to pseudomyotonia, muscular fasciculation, and myokymia [7] is the crucial element of the neuromyotonia.
It is unknown how frequent ARAN-NM is among the inherited neuropathies and the percentage of patients affected by inherited neuropathy caused by HINT1 mutations among patients with known cause of hereditary neuropathy.
We aimed to establish the importance of HINT1 mutations as the cause of hereditary neuropathy, in particular CMT2 and HMN among Czech patients.
Patients and methods
All tested patients signed informed consent for the CMT DNA diagnostics, and the study has been approved by the Central Ethical Committee of the University Hospital Motol.
Patients
Firstly, one hundred and sixty-six (166) unrelated patients with suspected hereditary neuropathy of axonal type/pure motor neuropathy (30 patients) or motor and sensory neuropathy (136 patients) were tested for mutations in the HINT1 gene by Sanger sequencing of all three coding exons and adjacent intronic regions. Most of the patients were previously tested for CMT1A/HNPP or other more frequent causes of CMT depending on their phenotype or provided clinical and electrophysiological information. No causal mutation was detected in these patients.
Patients with autosomal dominant inheritance were excluded. Sixteen patients had one affected sibling and healthy parents and were classified as autosomal recessive (AR). One hundred and fifty patients were sporadic cases.
Secondly, after sequencing of 166 patients, it was shown that the highly prevalent mutation is the p.R37P located in the exon 1 of the HINT1 gene. Thirty (30) unrelated patients were therefore tested by Sanger sequencing for only the first coding exon of the HINT1 gene. These patients were selected based on the same criteria as the patients above (CMT1A/HNPP and autosomal dominant inheritance excluded).
Thirdly, in order to test a large group of patients for the presence just of the prevalent mutation p.R37P, we selected 726 patients’ DNA from our database. These patients had unknown cause of the disease, and most of the patients were previously tested for CMT1A/HNPP or other more frequent causes of CMT depending on their phenotype or provided clinical and electrophysiological information. No causal mutation was detected in these patients.
Patients with autosomal dominant pedigree were excluded. These patients were tested with a TaqMan-based real-time PCR allelic assay.
Finally, patients with mutations in the HINT1 gene were contacted and invited to a neurogenetic consultation. The aim of this session was to explain the molecular genetic cause of the disease, to examine or re-examine the patients in detail, and help them to understand the specific features of this particular Charcot-Marie-Tooth disease subtype.
Methods
DNA sequencing by capillary electrophoresis
DNA was extracted from peripheral blood or saliva. All three exons and exon/intron boundaries of the HINT1 gene were polymerase chain reaction (PCR) amplified in three fragments. PCR primers are available upon request. PCR products were purified (FastSAP/ExoI, Thermo Scientific) and then sequenced using dye terminator reaction (BigDye Terminator v3.1, Applied Biosystems, Life Technologies). After dye terminator removal with Agencourt CleanSeq system (Beckman Coulter), the products were analyzed on an ABI 3130 automated genetic analyzer (Applied Biosystems, Life Technologies). Sequence traces were compared to a reference sequence NM_005340.5 using Sequencing Analysis Software (Applied Biosystems, Life Technologies) and Mutation Surveyor Software (SoftGenetics).
Allelic discrimination assay for mutation p.R37P
For this analysis, we used a Custom TaqMan® SNP Genotyping Assay manufactured by Applied Biosystems (Applied Biosystems, Life Technologies). A list of primers and probes is provided in online resource 1 (supporting information). Amplification and end-point analysis were performed on an ABI 7000 Sequence Detection System (Applied Biosystems, Life Technologies).
Further testing in families
For patients with HINT1 mutations, available family members were tested where possible, to prove the segregation of mutations in the family.
Genetic counselling and neurological examination
All patients with mutations in the HINT1 were offered genetic counselling and were examined by P.S. in order to avoid inter-examiner variability. Electromyography examinations were done by J.H. (paediatric patients) and R.M. (adult patients).
Results
Molecular genetic testing results
In the first group of patients, where all three exons of the HINT1 gene were tested, mutations were found in 11 unrelated patients (6.625 %). Among 30 patients originally classified as HMN, mutations were found in three of them (10 %). Among 136 patients originally classified as HMSN II, mutations were found in eight (5.88 %).
In the second group of patients, where only the first coding exon of the HINT1 was tested, mutations were found in three unrelated patients (10 %).
In the third group of patients, where only the presence of the p.R37P mutation was tested, mutation was found in altogether 10 patients (1.37 %). From these, five (5) patients were homozygotes for p.R37P mutation and five (5) patients carry the p.R37P mutation in a heterozygous state—on one allele only. These patients were tested by sequencing of all three coding exons. Mutation on the second allele was found only in one patient, the p.Q106X mutation.
Overall, mutations in the HINT1 gene are a surprisingly frequent cause of CMT among Czech patients.
Biallelic pathogenic mutations were found in 21 patients from 19 families. Four unrelated patients were found with mutation on one allele only, and therefore this result did not explain the cause of the CMT disease in these patients. These patients, heterozygotes, were re-evaluated. As for these patients, either the type of neuropathy, age of onset, or phenotype was very different from what has been observed in a group of patients with biallelic mutations. We therefore assumed these patients are heterozygotes for the p.R37P mutation only by chance and their neuropathy is caused by other causes. Four heterozygotes among 726 tested DNA allow us to estimate the p.R37P mutation heterozygote carrier frequency in the Czech population as being about 1:182.
Spectrum of mutations
Highly prevalent mutation was shown to be the p.R37P mutation (Figs. 1 and 2). Seventeen (17) unrelated patients carry the p.R37P mutation in a homozygous state. Two unrelated patients are compound heterozygotes for the p.R37P mutation and p.Q106X mutation. All patients with biallelic pathogenic mutations in the HINT1 gene carry the mutation p.R37P at least on one allele.
Detected mutations and their position relative to exons of the HINT1 gene. Blue box indicates exon, yellow box intron, red arrow mutations detected by Zimon et al., and green arrow mutations described in our study. (Part of this figure was inspired by the Alamut software, Interactive Biosoftware company)
Our report is the only cohort study on patients with HINT1 mutations following the molecular basis discovery report by Zimon et al. [5].
Clinical examination results
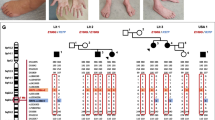
The clinical data of patients with HINT1 mutations are summarized in Tables 1 and 2. Patients are presented in Figs. 3, 4, 5, 6, and 7.
Electrophysiological study results are summarized in Table 2. The results of the electrophysiological studies of peripheral nerves (ENMG) significantly confirmed a primary axonal lesion of motor and sensory fibers. The conduction studies of motor and sensory fibers showed normal or nearly normal conduction velocities and decreased amplitude of CMAPs or SNAPs, respectively. No markers of demyelination (conduction slowing, temporal dispersion, or conduction block) were found.
The needle electromyography showed increased amplitude of motor unit action potentials (MUAPs) and reduction of recruitment pattern with temporal summation. We considered MUAP changes and recruitment pattern reduction as markers of chronic neurogenic lesion with reinnervation activity.
In some patients, neuromyotonic discharges in distal and proximal muscles were registered. The neuromyotonic discharges were provoked with a concentric needle electrode, and the frequency of discharges fluctuated only slightly from 50 to 80 Hz.
Our patients presented with early onset of symptoms (the end of the first life decade). Age at clinical diagnosis was between 13 and 55 years. Some of them were initially reported as HMN (or SMA III, patients younger at the time of clinical diagnosis). Some were initially reported as HMSN II or CMT2 (mostly patients of higher age at the time of diagnosis). Foot/toe extension plegia/weakness was present in almost all patients. On the other hand, foot deformity is not a constant feature. Some patients presented with muscle hypertrophy, esp. m.quadriceps. Regarding neuromyotonia, it was properly recognized only in three patients prior to molecular genetic diagnosis. However, when we actively searched for neuromyotonia signs, these were present in nine out of 11 patients re-examined after molecular clarification. One of them reported to have had noticed it in the past. Patients experienced difficulty in releasing grip and fingers after a strong voluntary hand contraction. For more detailed information and illustration of neuromyotonia, please see the electronic supplementary material no. 5 and no. 6.
Discussion
In the supplementary files of the paper by Zimon et al. [5], 50 patients are described with ARAN-NM. These patients presented with very similar symptoms. These patients are from Turkey, Serbia, Bulgaria, Croatia, Austria, Belgium, and Italy and also Roman ethnicity. In their cohort, action myotonia was present in 36/50 (72 %), which is in accordance with our results. A typical presenting sign was gait impairment at the end of the first life decade, similar to our observations.
Furthermore, there is a case report by Caetano et al. [9], in which a patient with a very similar phenotype is described. This patient was also initially tested by M. Zimon [5] and described in the report.
In addition, two patients with ARAN-NM, but with different mutations (p.C84R and p.H114R), are described in a paper by Zhao et al. [2]. However, there are no details about the phenotype. Their phenotype however seems somewhat different from our cohort. Those two patients presented with later onset of symptoms (17 and 28 years, respectively). No upper limb involvement is reported for these two patients. Both have had pes cavus, which is not a constant feature in our cohort. One of the patients described in that paper has hyporeflexia, one hyperreflexia. In contrast, our patients presented with absent/severely reduced L2–S2 reflexes.
Our report further expands and confirms the spectrum of phenotype of ARAN-NM. We present the largest cohort of patients with ARAN-NM, except for the supplementary files in the initial report by Zimon et al. [5].
As for the spectrum of mutations in the HINT1 gene, we have confirmed that the prevalent mutation is the p.R37P mutation (95.5 % of pathogenic alleles). We have also detected the p.Q106X mutation (in two unrelated patients) which has not been described before. The pathogenic character of this mutation is supported by its character (stop mutation) and occurrence (two unrelated families, three patients, always in combination with the prevalent p.R37P mutation on the other allele of the HINT1 gene). Targeted testing just for the p.R37P mutation and sequencing of the entire coding region of the HINT1 gene only in patients with the p.R37P on at least one allele is able to identify 100 % of all ARAN-NM patients in our population according to our observation. Therefore, testing will be targeted at first to p.R37P. This is a very cost-/labor-effective approach, simplifying DNA testing.
For comparison, eight different mutations in the HINT1 gene were detected in the study by Zimon et al. Fifty patients from 33 families were included, but the patients were recruited from several different populations though most of them from Serbia. Thirty-seven patients were homozygous for p.R37P. Six patients carry p.R37P in combination with other mutations on the second allele of the HINT1 gene. And seven patients carry two different mutations (different from p.R37P) on both alleles of the HINT1 gene.
Regarding the overall frequency of ARAN-NM in our population, mutations in the HINT1 gene are a surprisingly frequent cause of CMT2/HMN and patients with sporadic or recessive HMN or HMSN II should be tested for the presence of mutations in the HINT1 gene.
Our lab is the only facility in the Czech Republic offering and performing DNA testing for hereditary neuropathies. In summary, in our cohort of patients with inherited peripheral neuropathies (IPN), the molecular genetic cause has been identified in 1664 patients (884 unrelated families). From these, 726 patients (393 unrelated families) presented with CMT1A and 445 patients (229 unrelated families) have been diagnosed with HNPP. Four hundred and ninety-three (493) patients from 229 unrelated families carry point mutations in different genes associated with inherited peripheral neuropathy (IPN) of all types and inheritance patterns. Mutations in the GJB1 (Cx32) gene have been found in 172 patients (66 unrelated families). This accounts for 7.5 % of the whole IPN patient group with causal mutations.
There are 39 unrelated families (62 patients) in our database with causal mutations in the MPZ gene. This accounts for 4.4 % of the whole IPN patient group with causal mutations.
Overall, mutations have been identified in 27 unrelated families in the CMT2/HMN subgroup. Mutations in the BSCL2 gene (5 families, 0.6 % of the whole IPN group, 19 % of the HMN/CMT2 group), HSP22 (1 family, 4 % of the HMN/CMT2 group), GARS (1 family, 4 % of the HMN/CMT2 group), TRPV4 (1 family, 4 % of the HMN/CMT2 group), and HINT1 gene (19 families, 2.1 % of the whole group, 69 % of the HMN/CMT2 group) have been detected (Fig. 8).
Overall results for all patients with inherited peripheral neuropathy with HMN/CMT2 from our database where samples from all Czech hereditary neuropathy patients are collected. 1 indicates all IPN patients from our database with known mutation/molecular genetic diagnosis, 2 HMN/CMT2 patients with mutations in the BSCL2 gene (5 families), 3 HMN/CMT2 patients with mutations in the HSP22 gene (1 family), 4 HMN/CMT2 patients with mutations in the GARS gene (1 family), 5 HMN/CMT2 patients with mutations in the TRPV4 gene (1 family), and 6 HMN/CMT2 patients with mutations in the HINT1 gene (19 families)
For comparison, in our database, mutations have been identified in seventeen (17) unrelated families in the SH3TC2 gene (1.9 % of the whole IPN group), sixteen (16) unrelated families in the MFN2 gene (1.8 % of the whole IPN group), and seven (7) unrelated families in the GDAP1 gene (0.79 % of the whole IPN group). These data clearly present that HINT1 mutations are one of the most frequent causes of hereditary neuropathy—mainly CMT2/HMN in our population, more frequent than mutations in all the abovementioned genes. Moreover, mutations in the HINT1 gene are the fifth most frequent known cause of hereditary neuropathy in the Czech population with only CMT1A/HNPP and mutations in the GJB1 gene and the MPZ gene being more frequent.
Therefore, patients with CMT/HMN in other countries should be tested for ARAN-NM in order to establish the importance of HINT1 mutations as the cause of hereditary neuropathy outside the Czech Republic.
Summary
All patients experienced symptoms before the age of 9 years. Neuromyotonic discharges were noticed in original electrophysiological reports only in three patients with both pathogenic mutations despite a highly experienced neurologist and electrophysiologist. In general, the lower limbs were much more affected by distal weakness than the upper limbs. HINT1 mutations seem to be a surprisingly frequent cause of hereditary neuropathy (comparable to MFN2) even in patients without manifest neuromyotonia. All patients with CMT2 or HMN without a clear dominant transmission in the family should be tested for the prevalent mutation p.R37P in the first exon of the HINT1 gene.
References
Reilly MM, Murphy SM, Laura M (2011) Charcot-Marie-Tooth disease. J Peripher Nerv Syst 16(1):1–14. doi:10.1111/j.1529-8027.2011.00324.x
Zhao H, Race V, Matthijs G, De Jonghe P, Robberecht W, Lambrechts D, Van Damme P (2014) Exome sequencing reveals HINT1 mutations as a cause of distal hereditary motor neuropathy. Eur J Hum Genet 22(6):847–850. doi:10.1038/ejhg.2013.231
Rossor AM, Kalmar B, Greensmith L, Reilly MM (2012) The distal hereditary motor neuropathies. J Neurol Neurosurg Psychiatry 83(1):6–14. doi:10.1136/jnnp-2011-300952
Aminkeng F (2013) HINT1 mutations define a novel disease entity—autosomal recessive axonal neuropathy with neuromyotonia. Clin Genet 83(1):31–32. doi:10.1111/cge.12030
Zimon M, Baets J, Almeida-Souza L, De Vriendt E, Nikodinovic J, Parman Y, Battaloglu E, Matur Z, Guergueltcheva V, Tournev I, Auer-Grumbach M, De Rijk P, Petersen BS, Muller T, Fransen E, Van Damme P, Loscher WN, Barisic N, Mitrovic Z, Previtali SC, Topaloglu H, Bernert G, Beleza-Meireles A, Todorovic S, Savic-Pavicevic D, Ishpekova B, Lechner S, Peeters K, Ooms T, Hahn AF, Zuchner S, Timmerman V, Van Dijck P, Rasic VM, Janecke AR, De Jonghe P, Jordanova A (2012) Loss-of-function mutations in HINT1 cause axonal neuropathy with neuromyotonia. Nat Genet 44(10):1080–1083. doi:10.1038/ng.2406
Hahn AF, Parkes AW, Bolton CF, Stewart SA (1991) Neuromyotonia in hereditary motor neuropathy. J Neurol Neurosurg Psychiatry 54(3):230–235. doi:10.1136/jnnp.54.3.230
Lance JW, Durke D, Pollard J (1979) Neuromyotonia in the spinal form of Charcot-Marie-Tooth disease. Clin Exp Neurol 16:49–56
Vasilescu C, Alexianu M, Dan A (1984) Neuronal type of Charcot-Marie-Tooth disease with a syndrome of continuous motor unit activity. J Neurol Sci 63(1):11–25
Caetano JS, Costa C, Baets J, Zimon Phd M, Venancio Phd M, Saraiva Phd J, Negrao L, Fineza I (2014) Autosomal recessive axonal neuropathy with neuromyotonia: a rare entity. Pediatr Neurol 50(1):104–107. doi:10.1016/j.pediatrneurol.2013.08.028
Acknowledgments
We deeply thank the patients and their families for participating in the study. We thank all the referring clinical neurologists and clinical geneticist for sending patients for DNA testing to our laboratory. This study is supported by IGA MH CR No. NT 14348-3 and MH CZ—DRO, University Hospital Motol, Prague, Czech Republic 00064203.
Ethical standards
Experiments comply with the current laws of the country of origin.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Custom TaqMan® SNP Genotyping Assays designed by Applied Biosystems (AppliedBiosystem, LifeTechnologies). Sequences of Primers and TaqMan Probes. (PDF 10 kb)
Figure S2
Patient 11 at the age of 61. Calf muscle atrophy, foot plegia, genua recurvata. (GIF 675 kb)
Figure S3
Patient 17 at the age of 16. Muscles of hips and calves are evolved. Pedes planovalgi are present. Active foot extension is possible to 90°. (GIF 671 kb)
Figure S4
Patient 19 at the age of 14. She is able to squat, walk on toes, but unable to walk on heels. Steppage gait and foot extension plegia are present. (GIF 980 kb)
Neuromyotonia (MP4 5826 kb)
Neuromyotonia (MP4 2421 kb)
Rights and permissions
About this article
Cite this article
Laššuthová, P., Brožková, D.Š., Krůtová, M. et al. Mutations in HINT1 are one of the most frequent causes of hereditary neuropathy among Czech patients and neuromyotonia is rather an underdiagnosed symptom. Neurogenetics 16, 43–54 (2015). https://doi.org/10.1007/s10048-014-0427-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-014-0427-8