Abstract
To investigate the effect of recharge water temperature on bioclogging processes and mechanisms during seasonal managed aquifer recharge (MAR), two groups of laboratory percolation experiments were conducted: a winter test and a summer test. The temperatures were controlled at ~5±2 and ~15±3 °C, and the tests involved bacterial inoculums acquired from well water during March 2014 and August 2015, for the winter and summer tests, respectively. The results indicated that the sand columns clogged ~10 times faster in the summer test due to a 10-fold larger bacterial growth rate. The maximum concentrations of total extracellular polymeric substances (EPS) in the winter test were approximately twice those in the summer test, primarily caused by a ~200 μg/g sand increase of both loosely bound EPS (LB-EPS) and tightly bound EPS (TB-EPS). In the first half of the experimental period, the accumulation of bacteria cells and EPS production induced rapid bioclogging in both the winter and summer tests. Afterward, increasing bacterial growth dominated the bioclogging in the summer test, while the accumulation of LB-EPS led to further bioclogging in the winter test. The biological analysis determined that the dominant bacteria in experiments for both seasons were different and the bacterial community diversity was ~50% higher in the winter test than that for summer. The seasonal inoculums could lead to differences in the bacterial community structure and diversity, while recharge water temperature was considered to be a major factor influencing the bacterial growth rate and metabolism behavior during the seasonal bioclogging process.
Résumé
Afin d’étudier l’effet de la température de l’eau de recharge sur les processus et les mécanismes de colmatage biologique lors de la recharge artificielle saisonnière d’un aquifère (MAR), deux groupes de tests de percolation ont été menés en laboratoire : un test d’hiver et un test d’été. Les températures ont été contrôlées à environ ~5±2 °C et à environ ~15±3 °C, et les tests ont intégré l’introduction d’inoculum bactériens issus de l’eau du forage, prélevée en mars 2014 et en août 2015, respectivement pour les tests d’hiver et d’été. Les résultats indiquent que les colonnes de sable se colmatent ~10 fois plus rapidement lors du test d’été en raison d’un taux de croissance bactérienne 10 fois plus élevé. Les concentrations maximales de substances polymères extracellulaires (SPE) totales pour le test d’hiver ont été à peu près deux fois supérieures par rapport au test d’été, essentiellement dues à une augmentation d’environ 200 μg/g de sable, que ce soit pour les SPE faiblement liées et pour les SPE fortement liées. Dans la première moitié de la période d’expérimentation, l’accumulation de cellules bactériennes et la production de SPE ont induit un colmatage biologique rapide, que ce soit pour les tests d’hiver et pour ceux d’été. Par la suite, le développement bactérien a dominé le colmatage biologique lors du test d’été, tandis que l’accumulation de SPE faiblement liées a donné lieu à un colmatage biologique complémentaire lors du test d’hiver. Les analyses biologiques ont montré que les bactéries dominantes lors des expérimentations pour les deux saisons n’étaient pas les mêmes, et que la diversité des populations bactériennes était supérieure d’environ 50% en hiver, par rapport à l’été. Les inoculum saisonniers pourraient mener à des différences dans la structure et la diversité des communautés bactériennes, tandis que la température de l’eau de recharge a été considérée comme étant un facteur majeur d’influence du taux de croissance bactérienne et de leur métabolisme durant les processus de colmatage saisonnier.
Resumen
Se realizaron dos grupos de experimentos de percolación de laboratorio para investigar el efecto de la temperatura del agua de recarga en los procesos y mecanismos de obstrucción biológica durante la recarga de acuíferos con un manejo estacional (MAR): una prueba de invierno y una prueba de verano. Las temperaturas se controlaron a ~5±2 y ~15±3 °C, y las pruebas incluyeron inóculos bacterianos adquiridos en agua de pozo durante marzo de 2014 y agosto de 2015, para las pruebas de invierno y verano, respectivamente. Los resultados indicaron que las columnas de arena se obstruyeron ~10 veces más rápido en la prueba de verano debido a una tasa de crecimiento bacteriano 10 veces mayor. Las concentraciones máximas de sustancias poliméricas extracelulares totales (EPS) en la prueba de invierno fueron aproximadamente el doble que en la prueba de verano, causadas principalmente por un ~200 μg/g de aumento de arena tanto de EPS (LB-EPS) levemente ligada como de EPS fuertemente ligada (TB-EPS). En la primera mitad del período experimental, la acumulación de células bacterianas y la producción de EPS indujeron a una obstrucción biológica rápida en las pruebas de invierno y verano. Posteriormente, el aumento del crecimiento bacteriano dominó la obstrucción biológica en la prueba de verano, mientras que la acumulación de LB-EPS condujo a una mayor obstrucción biológica en la prueba de invierno. El análisis biológico determinó que las bacterias dominantes en los experimentos para ambas estaciones eran diferentes y que la diversidad de la comunidad bacteriana era ~50% más alta en la prueba de invierno que la del verano. Los inóculos estacionales podrían conducir a diferencias en la estructura y diversidad de la comunidad bacteriana, mientras que la temperatura del agua de recarga se consideró un factor principal que influye en la tasa de crecimiento bacteriano y el comportamiento del metabolismo durante el proceso de obstrucción biológica estacional.
摘要
本论文分别设计了冬季和夏季两组室内渗流试验,来研究回灌水温度对季节性人工回灌含水层微生物堵塞过程和机理。其中,冬季室内试验,回灌水温度为~5±2 °C,渗流试验细菌接种液取自现场回灌井(采样时间为2014年3月)。夏季室内试验,回灌水温度为~15±3°C,接种液取自同一回灌井(采样时间为2015年8月)。研究结果表明,夏季渗流试验中,含水介质中细菌生长速率比冬季试验细菌生长速率高10倍,砂柱堵塞速率比冬季砂柱堵塞速率快10倍。冬季试验中,细菌分泌的总胞外聚合物(Extracellular polymeric substances, EPS)浓度最大值是夏季试验中总EPS浓度最大值的2倍,这主要是由于冬季细菌分泌的紧密结合型EPS(Loosely bound EPS, LB-EPS)和松散结合型EPS (Tightly bound EPS, TB-EPS)浓度比夏季LB-EPS和TB-EPS浓度高~200 μg/g 砂样。在冬季和夏季渗流试验中,细菌细胞的积累和EPS的产生是导致砂柱前半阶段堵塞的主要原因。此后,细菌自身的生长是导致夏季试验砂柱堵塞的主要原因,而LB-EPS增加是冬季试验砂柱堵塞的主要原因。微生物学分析表明,冬、夏季试验砂柱中优势菌种种类不同,且冬季细菌群落多样性比夏季细菌群落多样性高50%。现场采集的接种液的季节性差异可能引起渗流试验中细菌群落结构和多样性不同。然而,回灌水温度的不同是导致季节性微生物堵塞过程中细菌生长速率和代谢行为差异的主要原因。
Resumo
Para investigar o efeito da temperatura da água de recarga nos processos e mecanismos de biocolmatação durante a recarga sazonal de um aquífero com gerenciamento de recarga artificial (GRA), dois grupos de experimentos laboratoriais de percolação foram realizados a fim de representar as estações de inverno e verão. As temperaturas foram controladas a ~5±2 e ~15±3 ºC, e os testes envolveram inóculos de bactérias adquiridos de água de poço durante março de 2014 e agosto de 2015, para os testes de inverno e verão, respectivamente. Os resultados indicaram que as colunas de areia entupiram ~10 vezes mais rápido no teste de verão devido a uma taxa de crescimento bacteriano 10 vezes maior. As concentrações máximas totais de substâncias poliméricas extracelulares (SPE) no teste de inverno foram aproximadamente duas vezes maiores do que no teste de verão, causado principalmente por um aumento de ~200 μg/g de areia, ambos em ligações fracas em SPE (LFc-SPE) e ligações fortes em SPE (LFt-SPE). Na primeira metade do período experimental, o acúmulo de células bacterianas e a produção de SPE induziram uma rápida biocolmatação nos testes de inverno e verão. Em seguida, o aumento do crescimento bacteriano dominou a biocolmatação no teste de verão, enquanto o acúmulo de LFc-SPE levou a uma posterior biocolmatação no teste de inverno. A análise biológica determinou que as bactérias dominantes nos experimentos de ambas as estações foram diferentes e que a diversidade da comunidade bacteriana foi ~50% maior no teste de inverno do que no verão. Os inóculos sazonais poderiam levar a diferenças na estrutura e diversidade da comunidade bacteriana, enquanto a temperatura da água de recarga foi considerada o maior fator influenciador sobre a taxa de crescimento bacteriano e sobre o comportamento do metabolismo durante o processo sazonal de biocolmatação.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Managed aquifer recharge (MAR) has been widely used to mitigate the worldwide water crisis, particularly in regions with water scarcity and uneven distribution of water resources (Dillon 2005; Kim et al. 2010). MAR has many advantages such as groundwater resource augmentation, seawater intrusion prevention, water storage, and water quality improvement through soil-aquifer treatment (Aoki et al. 2005). Virtually all engineered operations experience some degree of clogging that could markedly decrease the efficiency of injection wells by a drastic reduction in the saturated hydraulic conductivity (Ks) of the aquifer (Pavelic et al. 2011). Researchers have documented several causes of clogging, generally classified into physical, chemical and biological or a combination thereof (Hoffmann and Gunkel 2011; Pavelic et al. 2011). Physical mechanisms include sediment compaction (Pérez-Paricio and Carrera 2001), suspended solids in the recharge water infiltrating into the soil/sediments (Pavelic et al. 2011), and release of entrapped air bubbles (Rinck-Pfeiffer et al. 2000). Chemical clogging primarily results from precipitation of calcium carbonates (Baveye et al. 1998). Additionally, bioclogging can stem from the accumulation of bacterial cells and production of extracellular polymeric substances (EPS; Thullner 2010).
Previous studies that have focused on aquifer bioclogging mechanisms have demonstrated that the accumulation of bacterial cells in pore throats or at grain contacts causes a substantial reduction in the Ks of aquifer materials (Vandevivere and Baveye 1992). Studies have also reported that EPS produced by bacteria play a significant role in the clogging process. These exopolymers could affect Ks either by increasing the fluid viscosity or by decreasing the pore volume (Vandevivere and Baveye 1992). Xia et al. (2016) showed that two types of EPS—i.e., loosely bound EPS (LB-EPS) and tightly bound EPS (TB-EPS)—had different effects on Ks reduction in porous media and concluded that, compared to TB-EPS, LB-EPS led to a larger reduction in Ks during the bioclogging process. Additionally, poorly soluble gaseous compounds such as nitrogen and methane, can also cause bioclogging if the gas bubbles are sufficiently large to be trapped in the pore network (Kandra et al. 2015). Furthermore, other organisms such as fungi (Seki et al. 1996), have been observed in clogged porous media.
In most MAR systems, different water sources are used, e.g., river water, treated effluent, stormwater, desalinated seawater, rainwater, and even groundwater from other aquifers, depending on seasonal temperature variation. Temperature variation could cause a change in aquifer hydraulic conductivity because pore water viscosity is temperature dependent (Constantz 1998). Temperature also influences chemical reactions. Prommer and Stuyfzand (2005) demonstrated that, during managed recharge of aerobic water into an anaerobic aquifer, the oxidation of pyrite was temperature controlled. Additionally, temperature affects microbial growth and activity (Warren and Bekins 2015), hence the bioclogging in aquifers.
Previous studies have revealed that over 99% of bacteria present in subsurface environments cannot be cultured in the laboratory because artificial media fail to mimic completely the comprehensive abiotic and biotic conditions required for bacterial growth (Alain and Querellou 2009), and because bacteria cannot immediately adapt to the changes in conditions (Pham and Kim 2012). Therefore, culture-dependent bacterial populations likely underestimate the true diversity of the microbial community structure. Denaturing gradient gel electrophoresis (DGGE) of polymerase chain reaction (PCR)-amplified 16S rDNA gene fragments, a powerful molecular method, allows rapid detection of microbial community changes and provides accurate information regarding the distribution and composition of microbial species (Aydin et al. 2015). Combining culture-independent and culture-dependent methods to investigate bacterial communities offers an exact description and explanation of the bioclogging process in aquifers.
It is known that temperature can significantly affect microbial growth and activity (Warren and Bekins 2015); however, to the best of the authors’ knowledge, the quantitative effect of seasonal temperature variation on the bioclogging process and bacterial metabolism in aquifers has not been thoroughly elucidated to date. Therefore, this study attempts to investigate the effect of recharge water temperature on bacterial growth and metabolism and to understand the differences in the bioclogging process and temperature-variation-induced mechanisms in MAR systems by a laboratory culture experiment. In this study, a series of bacteria-inoculated sand columns, flowing with sterilized nutrient solution at different temperatures, were used in winter (i.e., March 2014) and summer (i.e., August 2015) to simulate the bioclogging process. The objectives of this work are: (1) to investigate the temporal variation of the relative saturated hydraulic conductivity (Ks′) in the sand columns during two seasons; (2) to investigate the bacterial growth and production of total EPS, LB-EPS and TB-EPS; (3) to compare the bioclogging process and the bacterial community in the two seasonal experiments (for a listing all terms and their definitions see Table 1).
.
Materials and methods
Porous media
Sand from a natural aquifer near the Dagu River (36.38024° N, 120.12089° E) in Jiaozhou city, Shandong province, China, was used as a model porous medium. After homogenizing, sieving and excluding the gravel fraction >2 mm, the sand was washed seven times with deionized Milli-Q water to remove clay or silt. Then, it was soaked in 0.25 M HCl for 24 h to remove trace metals, followed by treatment with 0.25 M NaOH and deionized Milli-Q water to adjust the pH to 7.0–7.2 (Yang et al. 2013). Between the acid and base treatment, the sand was washed using Milli-Q water until a neutral pH. Finally, it was baked at 550 °C for 2 h to remove organic matter according to a loss on ignition method (Qian et al. 2011). The particle size was analyzed according to the standard soil test method (Ministry of Water Resources of the People’s Republic of China 1999). The median diameter of the sand was 539 μm, and the particle-size-distribution curve is presented in Xia et al. (2016). The sand was analyzed for mineralogy using X-ray Diffraction (XRD, D/max-rB, Rigaku Co., Tokyo, Japan), and the result indicated it was dominated by quartz (86.4%), with minor compositions from feldspar (7.2%), dolomite (3.4%) and hornblende (3.0%).
Bacterial inoculum and nutrient solution
Bacterial inoculum was collected from a severely clogged recharge well near the Dagu River (36.38024° N, 120.12089° E) in March 2014 and August 2015, to closely simulate actual field aquifer bioclogging. The clogged recharge well was back washed by pumping at a rate of 833 L/min. Then, the discharge water was stored at 4 °C and immediately transported back to the laboratory. Before inoculation of each column, the discharge water was settled for 30 min, and the supernatant was used as the bacterial inoculum.
A synthetic nutrient solution containing glucose as the sole carbon source was used to stimulate bacterial growth. The nutrient solution consisted of (in milligrams per liter of deionized water): glucose, 58; NaCl, 5000 (0.5%, w/v); NH4+-N, 0.5; total phosphorous (TP), 0.01; MgSO4·7H2O, 45; CaCl2·2H2O, 20, and a trace elements solution, 1 ml/L (Xia et al. 2014a). The pH was adjusted to 7.0–7.2. Before being injected into the columns, the nutrient solution was autoclaved at 121 °C for 15 min and then oxygenated by fully stirring.
Column setup
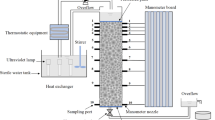
Acrylic glass columns, each 22-cm long with an inner diameter of 5 cm, were used for the percolation experiments (Fig. 1a). Piezometers were installed at 0, 2, 4, 6, 12, and 18 cm from the inlet (i.e., P1 in Fig. 1a) of the columns to monitor the hydraulic gradient. The columns were wet-packed with the prepared sand in small increments (~2 cm) using mild column vibration to minimize any layer heterogeneity or air entrapment. Fine stainless steel screens were placed at each end of the columns to prevent sand grains from creeping out. The packed columns were carefully sterilized via exposure to ultraviolet light under the wavelength of 254 nm. Parameters such as column volume, soil porosity and pore volume are presented in Table 2. Soil porosity was calculated based on the following equation (Danielson and Sutherland 1986),
where St represents soil porosity, ρb the bulk density, ρp the particle density. In the study, the bulk density (ρb) and particle density (ρp) were measured to be 1.63 and 2.65 g/cm3, according to the core method and pycnometer method (Blake and Hartge 1986), respectively.
For the inoculating procedure, the packed columns were injected downward with five pore volumes of the bacterial inoculum and then left for 12 h to foster cell attachment. Afterward, all the tubing and other parts in contact with the nutrient solution were replaced by sterilized ones to avoid contamination. A constant-temperature water bath (DC-0530, Baidian Yiqi Co., Ltd., Shanghai, China) was used to maintain the temperature of the sterilized nutrient solution (in the header tank). A peristaltic pump (BT100-2J, Longerpump Co., Ltd., Baoding, China) was used to pump the sterilized nutrient solution (in the header tank) to the constant head tank. The flow was vertically run from the top to the bottom of the inoculated columns under a constant hydraulic head loss. After flowing through the entire column, the nutrient solution was discharged to avoid column contamination.
Experimental design
Identical columns, arranged as eight duplicates, were involved in two seasonal percolation experiments: 16 columns representing winter (i.e., inoculums acquired in March 2014) and 16 columns representing summer (i.e., inoculums acquired in August 2015) (Fig. 1b); thus, there were 32 columns in total. All the sand columns were flushed with the sterilized nutrient solution for the predetermined times, namely, 0, 48, 96, 144, 192, 240, 288, or 336 h in March 2014 and 0, 4, 8, 10, 12, 14, 16, or 22 h in August 2015. Note, all the experimental conditions were identical except that the temperature of the nutrient solution was controlled at ~5±2 and ~15±3 °C in winter and summer, respectively. After flowing for the predetermined time, the flow rates and hydraulic head of the two parallel columns were measured to calculate the Ks and Ks′ values and then the columns were dismantled and sand samples were collected for biochemical analyses. Each sampling was performed in triplicate.
Saturated hydraulic conductivity
The saturated hydraulic conductivity Ks (cm/s) of the different layers of the sand columns could be calculated based on Darcy’s flow equation (Darcy 1856):
where the subscript i refers to the i-th layer from the inlet, delimited by two piezometers. For instance, i = 1 represents the piezometers installed at the inlet and the first 2 cm of the columns (i.e., P1 and P2 in Fig. 1a). In this equation, Q equals the flow rate (ml/min), A the cross-sectional area of the columns (cm2), (ΔH)i the hydraulic head difference of the i-th layer (cm), and Li the distance between the two piezometers at which the hydraulic head was measured (cm). In this study, the results are shown in terms of the relative saturated hydraulic conductivity Kis′ = Kis/Kis0, which is the ratio of the saturated hydraulic conductivity at the predetermined time t in comparison to its initial value.
Bacterial cell enumeration
After flowing for the predetermined time, the feed water valves of the two parallel columns were closed, and the excess nutrient solution drained off. Then, all the tubing and other parts in contact with the columns were carefully removed. The sand in the first layer of each column (i.e., the inlet to 2 cm) was collected and gently vortexed to mix. Then, weighed sand was sampled for bacterial cell enumeration, EPS quantification and DNA extraction.
Small, 1-g subsamples of the sand were weighed and transferred aseptically with a sterile spoon to a 10-ml sterile centrifuge tube. Each sampling was performed in triplicate. The samples were mixed with 5 ml of sterile phosphate buffer solution (PB, 10 mM, pH 7.4) and 0.5-g sterilized glass beads and then sonicated at 40 kHz, 30 W for 30 s. Then, the mixture was vortex oscillated at 4 °C for 1 min. The bacterial cell numbers was measured by serial diluting the suspension and plating it on a nutrient broth (NB) agar medium (Skill Bio Co., Ltd., Beijing, China): gelatin peptone (10 g), beef extract (3 g), sodium chloride (5 g) and agar (15 g) in 1-L deionized water at a final pH 7.0–7.2 (He et al. 2014). The plates were incubated at 37 °C for 72 h, and then visible bacterial colonies were manually counted. The data were reported as colony-forming units (CFUs) per gram of sand samples.
EPS extraction and quantification
A heating extraction method (Morgan et al. 1990) was modified to include a mild step for extracting LB-EPS and a harsh step for TB-EPS. After flowing for the predetermined time, the columns were carefully dismantled and sand from the first layer of each column (i.e., the inlet to 2 cm) was sampled in triplicate. In brief, a 5-g subsample of the sand was weighed and transferred aseptically with a sterile spoon to a 250-ml sterile plastic bottle. Then, the sand was mixed with 100-ml sterile PB solution and 2-g sterilized glass beads and treated by ultrasound at 40 kHz, 30 W for 30 s. After that, the suspension was harvested by centrifugation at 550 g for 15 min. Then, the supernatant was discharged, and the pellet was washed twice with the sterilized PB solution. Subsequently, the pellet was resuspended with 30-ml PB and centrifuged at 4,000 g and 4 °C for 15 min. The organic matter contained in the supernatant was considered to be LB-EPS. The pellet after LB-EPS extraction was further used for TB-EPS extraction. The residual pellet in the centrifuge tube was resuspended using 30-ml sterilized PB and then heated to 70 °C for 30 min in a water bath. After heating, the suspension was centrifuged at 4,000 g and 4 °C for 15 min. The organic matter in the supernatant was assumed to be TB-EPS.
After the extraction step, the organic matter was measured for polysaccharides and proteins contents. The polysaccharides (PSs, expressed in μg/g sand) were analyzed using a UV/VIS spectrophotometer (SQ2800, Unico, USA) according to the phenol-sulfuric acid method (Frølund et al. 1996) with a glucose standard (Kermel Chemical Reagent Co., Ltd., Tianjin, China). The proteins (PNs, expressed in μg/g sand) were assayed using the UV/VIS spectrophotometer according to the Bradford assay (Sharma and Babitch 1980) with albumin from bovine serum (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) as a standard. The sum of the PS and PN contents were assumed to be EPS.
Bacterial community analysis
DNA extraction
After flowing for 336 h in March 2014 and 22 h in August 2015, two parallel columns were carefully dismantled and sampling of each column was performed. Then 0.25-g subsamples of the sand in the first layer of each column (i.e., the inlet to 2 cm) were transferred aseptically with a sterile spoon to a 15-ml sterile centrifuge tube. Next, the samples were mixed with 10-ml sterile PB, and 0.5-g sterilized glass beads; these samples were then sonicated at 40 KHz, 30 W for 30 s. Next, the mixture was vortex oscillated at 4 °C for 1 min. Then, the suspension was transferred to a sterilized centrifuge tube and centrifuged at 550 g for 15 min. The supernatant was discarded, and DNA was extracted using the Power Soil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) following the manufacturer’s instructions. The pellet was transferred to the supplied Power Bead tube with 60 μl of the kit-supplied solution C1. The tube was incubated at 60 °C for 10 min and then shaken horizontally at maximum speed for 10 min using the MO BIO vortex adapter. The remaining steps were performed following the kit’s “Experienced user protocol.” The extracted DNA was stored at –80 °C until used.
PCR amplification and DGGE
The V3 region of the 16S rDNA gene was amplified by PCR using a universal bacterial primer set (forward primer, 5’-ATTACCGCGGCTGCTGG-3’; reverse primer, 5’-TGGCGGACGGGTGAGTAA-3’). PCR was performed in thin-walled tubes with a GeneAmp PCR system 9700 (Applied Biosystems, USA). One microliter of the template DNA was added to a reaction mix (final volume, 50 μl) containing dNTP (2.5 mM, 4 μl), 10 × PCR buffer (2.5 mM, 5 μl), forward and reverse primers (10 pmol, each 2 μl), Tap DNA polymerase (5 U/μl, 0.25 μl, TaKaRa, Dalian, China) and dd H2O (35.75 μl). The PCR reaction was performed as follows: (1) initial denaturation at 94 °C for 5 min; (2) 35 cycles of denaturation at 94 °C for 40 s, annealing at 55 °C for 40 s, and extension at 72 °C for 1 min; (3) final extension at 72 °C for 5 min. The DGGE of PCR products was carried out by using a D-Code System (Bio-Rad, USA). Gels were prepared and run in the following conditions: 8% (w/v) polyacrylamide, 1 × TAE buffer, linear gradient from 40 to 60% denaturant (where 100% denaturing agent was a mixture of 7-mol/L urea and 40% v/v deionized formamide). Electrophoresis was performed for 9 h at 150 V and 60 °C. After that, gels were stained with ethidium bromide and visualized with UV transilluminator (UVP, Inc., San Gabriel, CA, USA).
Analyses of DGGE spectra and bacterial diversity
DGGE banding patterns show the number and intensity of individual bands in each lane (Huang et al. 2012). Each DNA band represents a specific bacterial species in the community. The number of the bands directly reflects the genetic diversity of the bacterial community in samples. The higher the band number is, the more diverse the bacterial community is. In this study, Quantity One image analysis software (version 4.6.2, Bio-Rad Laboratories, Inc., USA) was used to quantify the number and intensity of the bands according to the manufacturer’s instructions.
The Shannon-Wiener index (H′) has been commonly used to characterize bacterial diversity in a community (Deng et al. 2012). In this study, the index was calculated based on the number and intensity values of the individual bands to compare the species richness and evenness of the dominated bacteria community in different seasonal experiments. The equations are as follows (Gong et al. 2012):
where H′ represents the Shannon-Wiener index, pi the relative abundance of the i-th species, Ni the intensity of band i, N the sum of the intensities of the bands in a lane, E the evenness of the dominated bacterial community, and S the richness, as determined from the number of the dominant bands in each lane.
Cloning, sequencing and phylogenetic analysis
The dominant bands in the DGGE gels were excised. Each excised piece was washed twice with 1 ml of sterilized deionized water and then placed into a 1.5-ml sterilized centrifuge tube to reclaim the DNA using SK1135 kit (Shanghai Sangon Biotech Co. Ltd.). The PCR products were sent to Sangon Biotech Co. Ltd. (Shanghai, China) for DNA sequencing. Then, the sequences were compared to the stored sequences in the National Center for Biotechnology Information (NCBI) database (NCBI 1988) using the BLAST algorithm. The homology of the bands was analyzed, and the neighbor-joining (NJ) method was used to establish a phylogenetic tree with the computer program MEGA version 6.0 (MEGA 1993).
Nucleotide accession numbers
The 17 nucleotide sequences were deposited in the GenBank database (NCBI 1988) under accession numbers KF680873-KF680878, KF680880, KF680882, KF680884, KF680887-KF680888, KX658645-KX658647, KF680877, and KX658648-KX658649.
Data sources
The experimental data for the winter test (i.e., March 2014) came from previous work (Xia et al. 2016), and data for the summer test were obtained from the percolation experiments conducted in August 2015. Several figures from Xia et al. (2016) are redrawn to show a better comparison between the two seasonal tests in this study.
Results and Discussion
In this study, only the first layer of the sand columns (i.e., the inlet to 2 cm) was investigated, as it was where the largest reduction in Ks occurred—see Tables S1 and S2 of the electronic supplementary material (ESM). This result was consistent with the previous findings (Xia et al. 2014b). Previous researchers revealed that the first 10% of the post-inlet length of the column is the principal bioclogging zone (Yang et al. 2013).
Temporal variation of the relative saturated hydraulic conductivity
The K1s′ value was used to compare normalized clogging of the first layer (i.e., the inlet to 2 cm) for all the sand columns (Fig. 2); note, Fig. 2, as in Figs. 3– 6, presents the results of one of the duplicate experiments to avoid cluttering the figures, whereby the data showed that, although certain deviations occurred due to porous media heterogeneity, the variation patterns were consistent (data for the duplicate experiments are presented in part II of the ESM). Figure 2 indicates that declines in K1s′ occurred in both the winter and summer experiments. In winter, a mild 5% reduction in K1s′ was observed within 48 h, followed by a sharp 94% decrease in the next 192 h. Toward the end of the experiment, the decreasing trend of K1s′ lessened again. In the whole test, the K1s′ was a 99.8% reduction to the initial value. In comparison, the summer test curve displayed a similar trend with a much faster reduction in K1s′. The K1s′ value slightly decreased by 5% over the initial 4 h, followed by a large 81% reduction over the next 10 h. At the end of the percolation test, the Ks′ was reduced to 1% of its initial value. While the overall behavior was similar, the sand columns were entirely clogged within 22 h in summer compared to 336 h in winter, which indicated that rapid clogging occurred in summer.
Bacterial growth and EPS production
Number of culturable bacteria
The variations in culturable bacteria of the first layer (i.e., the inlet to 2 cm) in the seasonal experiments are shown in Fig. 3. Although the culturable cell counts do not reflect the actual numbers of bacteria attached to the porous media, they are considered indirect estimates of both the abundance and activity of the attached bacteria. As shown in Fig. 3, the culturable cell numbers in both the winter and summer experiments reached maxima of ~×108 CFU/g sand, while the growth and decay periods in winter were significantly longer than those in summer. In the 336-h percolation experiment in winter, the numbers of culturable bacteria increased gradually within the initial 144 h, followed by a slightly decreasing trend in the next 192 h. The cell numbers reached a maximum value of 10.2 × 108 CFU/g sand, which corresponded to 500 times its initial value. Figure 3 also shows that the growth curve of the culturable bacteria was much shorter in summer compared to the experiment conducted in winter. After reaching a peak value of 5.4 × 108 CFU/g sand at 14 h, the total number of culturable bacteria gradually decreased as the percolation experiment progressed. This decrease implies that the higher temperature in the summer test (e.g., 15 °C) could accelerate bacterial growth rate and metabolic activity. This result agrees with Nagata et al. (2001), who concluded that the increasing temperature could favor microbial activity. Temperature governs biological metabolism by its effect on rates of biochemical reactions (Aledo and Jiménez-Riveres 2010); therefore, a favorable temperature could increase microbial activity (e.g., enzyme production, biodegradation) and membrane permeability. As the temperature decreases, cell membranes become increasingly viscous with decreasing membrane fluidity. The reduction in the fluidity of the membrane reduces transporter protein efficiency in the membrane and further influences transfer of oxygen and nutrition. This reduced efficiency finally leads to a decreased bacterial growth rate (Smet et al. 2015); however, under certain circumstances, the biochemical reaction rate may decline with increasing temperature (Aledo and Jiménez-Riveres 2010; Silverstein 2012). Aledo and Jiménez-Riveres (2010) demonstrated that there was a temperature switch point (e.g., ~30 °C for lactate dehydrogenase-B4a; ~20 °C for lactate dehydrogenase-B4b), above which the enzyme works slower due to the negative activation energy. Silverstein (2012) considered that this phenomenon occurred because of a dramatic conformational change of the enzyme (i.e., denaturation) in the temperature range of 20~30 °C.
Concentration and proportion of total EPS, LB-EPS and TB-EPS
Figure 4 demonstrates that there was a seasonal variation with generally increased LB-EPS, TB-EPS and total EPS in lower-temperature winter. In the winter test, the concentrations of LB-EPS and TB-EPS reached peak values of 540 and 155 μg/g sand, respectively, these values were much higher than those in summer (Fig. 4a,b). For the total EPS, the concentration increased sharply during the initial 192 h but fell slightly in the subsequent 144 h, with a peak value of 694 μg/g sand at 192 h in winter (Fig. 4c); however, total EPS reached a maximum of 417 μg/g sand within the initial 10 h in the summer experiment, which was lower than that in the winter experiment (Fig. 4c). This observation is in agreement with results reported by Wilén et al. (2008), who found that the total amount of EPS in flocs was significantly higher in the cold winter compared to the warm summer. Zhang et al. (2015) obtained negative correlations between the temperature and content of LB-EPS (R2 = −0.794, p < 0.01) and TB-EPS (R2 = −0.596, p < 0.01), which indicated that lower temperatures led to more LB-EPS and TB-EPS.
In this study, the reduced bacterial activity coupled with stimulated EPS production occurred in the winter test. This inconsistency may occur for two reasons. First, EPS production is a microbial response to their living environment (Li and Yang 2007). Microorganisms tend to produce more EPS to protect themselves from harsh environments such as low temperature or the presence of toxic compounds (Lyko et al. 2008). Although lower temperature inhibits a large number of bacteria, the surviving ones produce more exopolymers (Yang et al. 2013). Secondly, the seasonal differences in the proportion of polysaccharides and proteins in EPS may be responsible, as explained in the following section.
Figure 4d shows that the proportion of LB-EPS varied considerably in different seasons. The LB-EPS accounted for 31–80% of the total amount of EPS in winter, while they were the predominant component of the total EPS in summer (e.g., 91–95%). These results indicated that, compared to the TB-EPS, bacteria were inclined to secrete a significant amount of LB-EPS in a favorable living environment such as suitable temperature in summer; however, when encountering adverse conditions, bacteria altered their double-layer structure and tended to produce more TB-EPS. Compared to the LB-EPS, the TB-EPS were reported to be the primary surface by which cells protect against dewatering, low temperature, and nutrient shortage by strong interactions (Priester et al. 2006).
Concentration and proportion of major components in the total EPS
Polysaccharides and proteins are two major components of EPS (Chen et al. 2013; Han et al. 2017). Figure 5 shows the quantitative differences in polysaccharides and proteins in the total EPS in different seasonal experiments. It is apparent that the production rate of both proteins and polysaccharides was faster in summer than in winter, which caused the same characteristic of EPS variation. The proteins concentrations were similar in summer and winter, and both of the maxima reached ~100 μg/g sand (Fig. 5a). Zhang et al. (2015) noted that extracellular proteins secreted by bacteria were enzymes, and the slightly higher concentration in summer corresponded to the enhancement of microbial activity; however, the concentration of exopolysaccharides in the winter, which reached ~600 μg/g sand, was nearly twice that in the summer (~320 μg/g; Fig. 5b). This difference is likely because the polysaccharides, which are highly biodegradable polymers, are more likely to be easily degraded in the presence of high exoenzymes in the summertime (Zhang et al. 2015).
Figure 5c shows further differences in the proportions of polysaccharides to total EPS. Proportions in both the summer and the winter are above ~58%, especially in the winter, when they are over 90% on average. This result indicates that the polysaccharides are dominated by the production of total EPS. During the early stage of biofilm formation, bacterial cells tend to produce more exopolysaccharides than other exopolymers, which would promote cell adhesion onto the surfaces of porous media (Lopes et al. 2000). According to Vandevivere and Kirchman (1993), the polysaccharides/proteins ratio could be five-fold greater for attached cells than for free-living cells. Bacteria mediate cell cohesion and adhesion and maintain the structural integrity of the biofilm matrix by the production of exopolysaccharides (Tsuneda et al. 2001); therefore, polysaccharides are predominant in total EPS as the experimental results show.
In the winter test, the proportions of polysaccharides to total EPS were approximately 82–96%. The accumulation of the polysaccharides led to a significant amount of the total EPS. However, the proportions in the summer test (58–85%) had a ~20% reduction relative to those in the winter (82–96%). This difference may occur because more proteins were secreted during the period of increasing microbial activity, and the polysaccharides were biodegraded in the presence of exoenzymes (i.e., proteins) in the summer (Zhang et al. 2015).
Seasonal variations in the bioclogging process of the sand columns
The variations in the bacterial cell numbers, TB-EPS and LB-EPS concentrations versus the relative saturated hydraulic conductivities in the two seasonal experiments are shown in Fig. 6. The bioclogging processes were divided into four stages, regardless of the experiment season. In winter (Fig. 6a–c), the bacterial cell numbers and the two EPS concentrations changed with the decreasing relative saturated hydraulic conductivities. During stage I (i.e., 0–48 h), the K1s′ value decreased by 5%, with the bacterial cell numbers increasing from 6.39 to 7.10 log10 CFU/g sand, while the concentrations of TB-EPS and LB-EPS had almost no change. This outcome implies that the initial decline in K1s′ resulted from the bacteria growth. For stage II (i.e., 48–144 h), the K1s′ decreased by 67%, with increased in both the cell numbers and the two EPS concentrations. However, during stage III (i.e., 144–192 h), as the bacterial cell numbers and the TB-EPS began to decline after reaching their maxima, the concentration of LB-EPS further increased from 314 to 540 μg/g sand, which could be responsible for the further decrease in K1s′ (17%). At stage IV (i.e., after 192 h), complete clogging occurred with decreases of all three factors (the cell numbers, the TB-EPS and the LB-EPS), suggesting there might be other unknown influences as well.
The summer bacterial cell numbers were not substantially different to those in winter. However, compared to winter, the EPS concentrations in summer changed significantly, especially the TB-EPS, which was relatively stable and close to zero during the summer bioclogging process. The differences in the clogging process between summer and winter mainly occurred in stage III (Fig. 6d–f). At stage I (i.e., 0–4 h), the K1s′ decreased by 8% due to the increasing cell numbers from 6.15 to 6.82 log10 CFU/g sand, even with no obvious rise of the TB-EPS or the LB-EPS. During stage II (i.e., 4–10 h), the growth of bacterial cells and production of LB-EPS led to a 39% reduction in K1s′, with no obvious increase of the TB-EPS, a result different from the pattern in the winter test. Then, a continuous reduction in K1s′ was observed at stage III (i.e., 10–14 h), with the cell numbers increasing from 7.63 to 8.73 log10 CFU/g sand. There was no increase in the TB-EPS or the LB-EPS, again somewhat different from the winter results. Correspondingly, the K1s′ value further decreased by 10% due to other unexplained reasons in stage IV (i.e., after 14 h).
The results demonstrate that, although the variation patterns were similar in winter and summer during stages I and II of the bioclogging process, they were somewhat different in stage III. The major contributors in stage III were the bacterial cell numbers in summer and LB-EPS in winter. In the summer experiment, the concentration of TB-EPS leveled off to an almost constant value during the entire bioclogging process and contributed little to the reduction of K1s′, which was also different from the situation in the winter test.
Seasonal variations in the bacterial community
The bacterial community structure
The intensity of the DGGE bands can indicate the abundance of the bacterial community (Huang et al. 2012). Figure 7 and Table 3 both show that there were some differences in the bacterial community structure between the two seasonal experiments. Figure 7a demonstrates that the dominant bacteria in the winter experiment were at bands 3, 4, 5, 7 and 9, which corresponded to Janthinobacterium sp., Rahnella aquatilis, Tolumonas auensis, Acidovorax radicis and Aquabacterium commune, respectively (Table 3). By contrast, four predominant bands (i.e., bands 12, 15, 16 and 17) were present in the summer test (Fig. 7b), corresponding to Brevundimonas diminuta, T. auensis and Acidovorax radicis (Table 3). The taxonomic assignment was also confirmed using the phylogenetic tree in Fig. 8. By phyla taxonomic level, Proteobacteria dominated the bacterial community in both experiments, with the average relative abundances of 91.6% in winter and 84.2% in summer, respectively (data presented in Tables S3 and S4 of the ESM). At the level of class, Betaproteobacteria accounted for 51.4% of the classes presented in the winter experiment, following by Gammaproteobacteria, Alphaproteobacteria and Deltaproteobacteria, with relatively low proportions of 26.5, 7.3 and 6.7%, respectively. Similarly, Betaproteobacteria was the dominant class in the summer experiment with the proportion of 37.9%, following by Gammaproteobacteria and Alphaproteobacteria, which accounted for 30.9 and 15.7%, respectively (data presented in Tables S3 and S4 of the ESM).
Note that several dominant bacteria, including Rahnella aquatilis, T. auensis in winter and Brevundimonas diminuta, Tolumonas auensis in summer, were anaerobic gram-negative bacteria and primarily presented in urine. This implies that the clogging induced bacteria that originated from anthropogenic sources, since the sampling site was located downstream of the Dagu River, which flows through many cities (e.g., Zhaoyuan, Laixi, Pingdu, etc.) and has been severely impacted by human activities (Zhang and Xu 2009).
Diversity of the bacterial community
The diversity of the bacterial community in the seasonal experiments was analyzed based on the DGGE analysis of the amplified partial 16S rDNA genes. The Shannon-Wiener index (H′) provides the basic information on the shift in the bacterial community structure in regard to bacterial diversity. Table 4 shows a 55% increase of H′ in winter, which indicates a higher bacterial diversity under the colder temperature; meanwhile, the winter samples showed a larger number of dominant bands (S score) and higher evenness (E score) compared to summer. It was expected that although the favorable summer temperature enabled drastic short-term bacterial growth (Fig. 3), it would lead to a less diverse and less evenly distributed bacterial community structure. An increase of bacterial diversity was observed in the winter experiment, which revealed that the low temperature tended to promote diversity and a more even distribution of the bacterial community.
Implication of the seasonal bacterial community
The results indicated that some differences occurred in the bacterial community structure and diversity during the winter and summer experiments, particularly the dominant bacteria. The differences may stem from two possible reasons. On the one hand, the bacterial composition in the seasonal inoculums could be different due to in situ sampling, which were induced by complex field conditions (e.g. temperature, recharge water, etc.). On the other hand, a dynamic shift in bacterial community structure could occur due to changes in recharge water temperature during the bioclogging process in the seasonal experiments. Since the temperature-induced change of the bacterial community structure is a long-term process (He et al. 2014), it is inferred that the differences in the bacterial community in the clogged columns primarily resulted from the former reason.
The differences in the seasonal inoculums might also have a potential influence on the bacterial growth and EPS production; however, the influence is inferred to be weaker than that of temperature. Figure 6 shows that the bacterial growth pattern and the bacterial cell numbers were similar in the two seasonal experiments, while the difference in the growth rates was a multiple of ten. As discussed in section ‘Number of culturable bacteria’, the higher temperature could significantly accelerate metabolic activity and induce the much faster bacterial growth rate in the summer test. In addition, the dominant bacteria in both experiments had the ability to produce high amount of EPS (e.g., Badireddy et al. 2008; Douterelo et al. 2013; Liao et al. 2017), whereas EPS in summer were much lower than those in winter especially for the TB-EPS. It also implies that the differences that occurred in EPS production might be mainly induced by recharge water temperature. The aforementioned qualitative analysis inferred that the temperature could have a dominant influence on the bacterial growth and EPS production. However, to quantify the respective influences of temperature and the bacterial community, there needs to be further controlled experiments, which is the goal of the authors’ future research.
Conclusions
A series of bacterial inoculated sand columns flowing with sterilized nutrient solution at different temperatures was performed in March 2014 and August 2015 to simulate bioclogging processes during seasonal MAR. Based on the comparison of the bioclogging process in both the winter and summer experiments, the present study confirmed that, during seasonal MAR, the recharge water temperature could significantly affect the bacterial growth and metabolism behavior, and further induce differences in the bioclogging rate and process. The sand column clogging was ~10 times faster in summer than in winter, because of a faster culturable bacteria growth rate and EPS production in the favorable summer temperature. However, the maximum concentrations of total EPS in winter were about twice those of summer, a phenomenon that is primarily caused by a ~200 μg/g sand increase of both LB-EPS and TB-EPS. Although the low temperature decreased the bacterial growth rate, the surviving bacteria could increase production of exopolymers in order to protect them from harsh environments. The TB-EPS concentration remained low (<50 μg/g sand) during the entire bioclogging process in summer and contributed little to the total EPS (<10%). For the bioclogging processes, the behaviors were similar in stages I and II in both summer and winter, and the differences mainly occurred in stage III. In the third stage, the major contributor to the reduction of the relative saturated hydraulic conductivity was bacterial cell numbers in summer, while the LB-EPS led to the decreasing Ks′ in winter. The dominant bacteria in the winter experiment were T. auensis, Acidovorax radices, Janthinobacterium sp., Rahnella aquatilis, and Aquabacterium commune, while T. auensis, Acidovorax radices, and Brevundimonas diminuta dominated in summer. For the bacterial community diversity, a ~50% increase occurred in the winter experiment. The differences in the bacterial community structure and diversity could mainly result from the seasonal inoculums. Although the different inoculums could also have an impact on the bacterial growth rate and metabolism behavior, the recharge water temperature was considered to be the dominant factor in the seasonal bioclogging process. Quantitative study on contributions of inoculum and recharge water temperature on seasonal managed aquifer recharge will be undertaken in the future research.
References
Alain K, Querellou J (2009) Cultivating the uncultured: limits, advances and future challenges. Extremophiles 13:583–594
Aledo CJ, Jiménez-Riveres S (2010) The effect of temperature on the enzyme-catalyzed reaction: insights from thermodynamics. J Chem Educ 87:296–298
Aoki C, Memon MA, Mabuchi H (2005) Water and wastewater reuse: an environmentally sound approach for sustainable urban water management. UNEP, Nairobi, Kenya
Aydin S, Shahi A, Ozbayram EG, Ince B, Ince O (2015) Use of PCR-DGGE based molecular methods to assessment of microbial diversity during anaerobic treatment of antibiotic combinations. Biores Technol 192:735–740
Badireddy AR, Chellam S, Yanina S, Gassman P, Rosso KM (2008) Bismuth dimercaptopropanol (BisBAL) inhibits the expression of extracellular polysaccharides and proteins by Brevundimonas diminuta: implications for membrane microfiltration. Biotech Bioeng 99(3):634–643
Baer ML, Ravel J, Chun J, Hill RT, Williams HN (2000) A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int J Syst Evol Microbiol 50:219–224
Baveye P, Vandevivere P, Hoyle BL, DeLeo PC, Lozada DS (1998) Environmental impact and mechanisms of the biological clogging of saturated soils and aquifer materials. Crit Rev Env Sci Tec 28(2):123–191
Blake GR, Hartge KH (1986) Particle density. In: Methods of soil analysis, part 1: physical and mineralogical methods. American Society of Agronomy, Madison, WI, pp 377–382
Bouvet PJM, Grimont PAD (1986) Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended description of Ainetobacter calcoaceticus and Acinetobacter lwoffii. Int J Evol Microbiol 36:228–240
Chen YP, Zhang P, Guo JS, Fang F, Gao X, Li C (2013) Functional groups characteristics of EPS in biofilm growing on different carriers. Chemosphere 92(6):633–638
Constantz J (1998) Interaction between stream temperature, stream flow, and groundwater exchanges in alpine streams. Water Resour Res 34(7):1609–1616
Danielson RE, Sutherland PL (1986) Porosity. In: Methods of soil analysis, part 1: physical and mineralogical methods. American Society of Agronomy, Madison, WI, pp 443–461
Darcy H (1856) Les fontaines publications de la ville de Dijon [The fountains publication of the city of Dijon]. Dalmont, Paris
De Ley J, Segers P, Gillis M (1978) Intra and intergeneric similarities of Chromobacterium and Janthinobacterium ribosomal ribonucleic acid cistrons. Int J Syst Bacteriol 28:154–168
Deng B, Shen CH, Shan XH, Ao ZH, Zhao JS, Shen XJ, Huang ZG (2012) PCR-DGGE analysis on microbial communities in pit mud of cellars used for different periods of time. J Inst Brew 118: 120-126
Dillon P (2005) Future management of aquifer recharge. Hydrogeol J 13:313–316
Douterelo I, Sharpe RL, Boxall JB (2013) Influence of hydraulic regimes on bacterial community structure and composition in an experimental drinking water distribution system. Water Res 47(2):503–516
Fischer-Romero C, Tindall BJ, Jüttner F (1996) Tolumonas auensis gen. nov., sp. nov., a toluene-producing bacterium from anoxic sediments of a freshwater lake. Int J Syst Bacteriol 41(1):183–188
Frølund B, Palmgren R, Keiding K, Nielsen PH (1996) Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res 30(8):1749–1758
Gong MG, Tang M, Zhang QM, Feng X (2012) Effects of climatic and edaphic factors on arbuscular mycorrhizal fungi in the rhizosphere of Hippophae rhamnoides in the Loess Plateau, China. Acta Ecolog Sin 32:62–67
Han ZZ, Meng RR, Yan HX, Zhao H, Han M, Zhao YY, Sun B, Sun YB, Wang J, Zhuang DX, Li WJ, Lu LX (2017) Calcium carbonate precipitation by Synechocystis sp. PCC6803 at different Mg/Ca molar ratios under the laboratory condition. Carbonates Evaporites 32(4):561–575
He G, Yi F, Zhou S, Lin J (2014) Microbial activity and community structure in two terrace-type wetlands constructed for the treatment of domestic wastewater. Ecol Eng 67:198–205
Hoffmann A, Gunkel G (2011) Bank filtration in the sandy littoral zone of Lake Tegel (Berlin): structure and dynamics of the biological active filter zone and clogging processes. Limnologica 44(1):10–19
Huang T, Wei W, Su J, Zhang H, Li N (2012) Denitrification performance and microbial community structure of a combined WLA-OBCO system. PloS One 7(11):e48339
Izard D, Gavini F, Trinel PA, Leclerc H (1979) Rahnella aquatilis, nouveau member de la famille des Enterobacteriaceae. Ann Microbiol 130A:163–177
Kalmbach S, Manz W, Wecke J, Szewzyk U (1999) Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov. and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int J Syst Bacteriol 49:769–777
Kandra HS, Asce SM, Callaghan J, Deletic A, McCarthy DT (2015) Biologicla clogging in storm water filters. J Environ Eng 141(2):1–8
Kato Y, Asahara M, Goto K, Kasai H, Yokota A (2008) Methylobacterium persicinum sp. nov., Methylobacterium komagatae sp. nov., Methylobacterium brachiatum sp. nov., Methylobacterium tardum sp. nov. and Methylobacterium gregans sp. nov., isolated from freshwater. Int J Syst Evol Microbiol 58:1134–1141
Kim JW, Choi H, Pachepsky YA (2010) Biofilm morphology as related to the porous media clogging. Water Res 44:1193–1201
Li D, Rothballer M, Schmid M, Esperschütz J, Hartmann A (2011) Acidovorax radicis sp. Nov., a wheat-root-colonizing bacterium. Int J Syst Evol Microbiol 61:2589–2594
Li XY, Yang SF (2007) Influence of extracellular polymeric substances (EPS) on the flocculation, sedimentation and dewaterability of activated sludge. Water Res 41:1022–1030
Li Y, Kawamura Y, Fujiwara N, Naka T, Liu H, Huang X, Kobayashi K, Ezaki T (2004) Rothia aeria sp. Nov., Rhodococcus baikonurensis sp. Nov. and Arthrobacter russicus sp. Nov., isolated from air in the Russian space laboratory. Mir Int J Syst Evol Microbiol 54:827–835
Liao J, Fang C, Yu J, Sathyagal A, Willman E, Liu WT (2017) Direct treatment of high-strength soft drink wastewater using a down-flow hanging sponge reactor: performance and microbial community dynamics. Appl Microbiol Biot 101(14):5925–5936
Lopes FA, Vieira MJ, Melo LF (2000) Chemical composition and activity of a biofilm during the start-up of an airlift reactor. Water Sci Technol 41:105–111
Lyko S, Wintgens T, Al-Halbouni D, Baumgarten S, Tacke D, Drensia K (2008) Long-term monitoring of a full-scale municipal membrane bioreactor-characterisation of foulants and operational performance. J Membr Sci 317:78–87
MEGA (1993) Molecular evolutionary genetics analysis. http://www.megasoftware.net/home/download. Accessed March 2018
Ministry of Water Resources of the People’s Republic of China (1999) Standard for Soil Test Method, SL 237–1999 (in Chinese). Water Power Process, Beijing
Morgan JW, Forster CF, Evison L (1990) A comparative study of the nature of biopolymers extracted from anaerobic and activated sludges. Water Res 24(6):743–750
Nagata T, Fukuda R, Fukuda H, Koike I (2001) Basin-scale geographic patterns of bacterioplankton biomass and production in the subarctic Pacific, July–September 1997. J Oceanogr 57:301–313
NCBI (1988) National Center for Biotechnology Information, US National Library of Medicine. https://www.ncbi.nlm.nih.gov/. Accessed March 2018
Pavelic P, Dillon PJ, Mucha M, Nakai T, Barry KE, Bestland E (2011) Laboratory assessment of factors affecting soil clogging of soil aquifer treatment systems. Water Res 45:3153–3163
Pérez-Paricio A, Carrera J (2001) Clogging and heterogeneity. In: Jensen KH (ed) Artificial recharge of groundwater. Final report, EC project ENV4-CT95-0071, Luxembourg 92-894-0186-9, EC, Brussels, pp 51–58
Pham VHT, Kim J (2012) Cultivation of unculturable soil bacteria. Trends Biotechnol 30(9):475–484
Priester JH, Olson SG, Webb SM, Neu MP, Hersman LE, Holden PA (2006) Enhanced exopolymer production and chromium stabilization in Pseudomonas putida unsaturated biofilms. Appl Environ Microbiol 72:1988–1996
Prommer H, Stuyfzand PJ (2005) Identification of temperature-dependent water quality changes during a deep well injection experiment in a pyritic aquifer. Environ Sci Technol 39:2200–2209
Qian B, Liu L, Xiao L (2011) Comparative tests on different methods for content of soil organic matter. J Hohai University (Natural Sciences) 39(1):34–38
Rinck-Pfeiffer S, Ragusa S, Sztajnbok P, Vandevelde T (2000) Interrelationships between bioclogging, chemical, and physical processes as an analog to clogging in aquifer storage and recovery (ASR) wells. Water Res 34(7):2110–2118
Schleifer KH, Kloos WE (1975) Isolation and characterization of Staphylococci from human skin I. Amended description of Staphylococcus epidermidis and Staphylococcus saprophyticus and description of three new species: Staphylocyccus cohnii, Staphylococcus haemolyticus, and Staphylococcus sylosus. Int J Syst Bacteriol 25(1):50–61
Segers P, Vancanneyt M, Pot B, Torck U, Hoste B, Dewettinck D, Falsen E, Kersters K, De Voss P (1994) Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Büsing, Döll, and Freytag 1953 in Brevundimonas gen. ov. as Brevundimonas diminuta comb. nov., respectively. Int J Syst Bacteriol 44(3):499–510
Seki K, Miyazaki T, Nakano M (1996) Reduction of hydraulic conductivity due to microbial effects. Trans Jpn Soc Irrig Drain Reclam Eng 181:137–144
Sharma SK, Babitch JA (1980) Application of Bradford’s protein assay to chick brain subcellular fractions. J Biochem Bioph Meth 2(4):247–250
Silverstein TP (2012) Falling enzyme activity as temperature rises: negative activation energy or denaturation? J Chem Educ 89:1097–1099
Smet C, Derlinden EV, Mertens L, Noriega E, Impe JFV (2015) Effect of cell immobilization on the growth dynamics of Salmonella typhimurium and Escherichia coli at suboptimal temperatures. Int J Food Microbiol 208:75–83
Thullner M (2010) Comparison of bioclogging effects in saturated porous media within one- and two-dimensional flow systems. Ecol Eng 36:176–196
Tsuneda S, Park S, Hayashi H, Jung J, Hirata A (2001) Enhancement of nitrifying biofilm formation using selected EPS produced by heterotrophic bacteria. Water Sci Technol 43:197–204
Vandevivere P, Baveye P (1992) Effect of bacterial extracellular polymers on the saturated hydraulic conductivity of sand columns. Appl Environ Microbiol 58(5):1690–1698
Vandevivere P, Kirchman DL (1993) Attachment stimulates exopolysaccharide synthesis by a bacterium. Appl Environ Microbiol 59:3280–3286
Warren E, Bekins BA (2015) Relating subsurface temperature changes to microbial activity at a crude oil-contaminated site. J Contam Hydrol 182:183–193
Wilén BM, Lumley D, Mattsson A, Mino T (2008) Relationship between floc composition and flocculation and setting properties studies at a full scale activated sludge plant. Water Res 42:4404–4418
Xia L, Zheng XL, Duan YH, Peng T (2014b) Analysis of process and mechanism of bioclogging in aqueous media. J Hydraul Eng 45(6):749–754
Xia L, Zheng XL, Shao HB, Xin J, Peng T (2014a) Influences of environmental factors on bacterial polymeric substances production in porous media. J Hydrol 519:3153–3162
Xia L, Zheng XL, Shao HB, Xin J, Sun ZY, Wang LY (2016) Effects of bacterial cells and two types of extracellular polymers on bioclogging of sand columns. J Hydrol 535:293–300
Yang SY, Ngwenya BT, Butler IB, Kurlanda H, Elphick SC (2013) Coupled interactions between metals and bacterial biofilms in porous media: implications for biofilm stability, fluid flow and metal transport. Chem Geol 337-338:20–29
Zhang JW, Xu HK (2009) Dagu River area source pollution of agricultural fertilizer and its countermeasure. J Anhui Agri Sci 37(16):7632–7633 7657
Zhang WJ, Yang P, Xiao P, Xu SW, Liu YY, Liu F, Wang DS (2015) Dynamic variation in physicochemical properties of activated sludge floc from different WWTPs and its influence on sludge dewaterability and settleability. Colloid Surface A 467:124–134
Acknowledgements
Sincere gratitude is expressed to the reviewers for their many helpful comments and constructive criticisms.
Funding
The authors would like to thank the National Science Foundation of China (41641020), Scientific Research Foundation of Shandong University of Science and Technology for Recruited Talents (2017RCJJ032), the China Postdoctoral Science Foundation (2016M592219), and the Qingdao Postdoctoral Applied Research Project (2015195) for their financial support. Xilai Zheng was also financially supported by the National Key Research Project (2016YFC0402810).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 1235 kb)
Rights and permissions
About this article
Cite this article
Xia, L., Gao, Z., Zheng, X. et al. Impact of recharge water temperature on bioclogging during managed aquifer recharge: a laboratory study. Hydrogeol J 26, 2173–2187 (2018). https://doi.org/10.1007/s10040-018-1766-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10040-018-1766-6