Abstract
The relevance of oligodendroglial histological features to patient prognoses is controversial. 93 LrGGs resected for about 2 decades were re-assessed based on WHO2007 with special interest to pure oligodendroglial diagnosis (oligodendroglioma or anaplastic oligodendroglioma) and presence of CFO features. Those histological features, patients OS, and tumor chromosomal/genetic characteristics were correlated each other in each of the 3 IDH-1p/19q-based molecular groups. There was significant association between 1p19q status with the oligodendroglial histological diagnosis as well as presence of CFO in the entire cohort. The oligodendroglial diagnosis was associated with longer OS in IDHmut/codel group; however, this association was not significant in the multivariate analyses. In IDHmut/noncodel and IDH-wildtype groups, the oligodendroglial diagnosis was not associated with patient OS. Presence of CFO was not associated with patient OS in any molecular groups. Gain of 8q was associated with the oligodendroglial diagnosis in IDHmut/noncodel group. Neither the oligodendroglial diagnosis nor CFO was predictive for the methylation status of the MGMT gene in any molecular groups. The oligodendroglial histological features are not independently predictive of either patient prognosis or chemotherapeutic response in LrGGs, leaving the possibility of marginal favorable association only in IDHmut/codel tumors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Based on the accumulating evidence that molecular characteristics is more closely associated with clinical outcomes as compared with histological classification in many tumor types, the revised WHO classification of tumors of the central nervous system in 2016 [1] incorporated the well-established molecular factors as well as histological features, and the final diagnosis would be the integrated diagnosis considering both of them. Indeed, the diffuse gliomas of grade II and III (lower-grade gliomas, LrGGs) are now classified into 3 molecular subgroups, virtually regardless of histological features, based on IDH mutation status and presence or absence of 1p19q codeletion. On the other hand, the significance of histological features independent of IDH-1p19q status, especially oligodendroglial features, remained controversial, and the oligodendroglial morphology itself might be prognostic for patients with LrGGs [6, 22].
To reappraise the significance of oligodendroglial morphology in terms of clinical outcome, and to investigate molecular background possibly associated with oligodendroglial morphology, we studied consecutive cases of LrGG for about 2 decades treated in a single institute. The tumors were histologically re-assessed based on WHO2007 [2] (the most recent one solely based on histology), and oligodendroglial morphology was correlated with patient OS and molecular characteristics including promoter methylation of the MGMT gene, copy number aberration (CNA) as assessed by CGH, mutations in the IDH, ATRX, and TERT genes in each of the IDH-1p19q based 3 molecular groups. Prognostic relevance of those molecular-genetic factors was also investigated. The NGS-based cancer gene panel test was performed for IDHmut/codel tumors, and DNA methylome-based classification was performed for IDHwild tumors.
Materials and methods
Patients and tissue samples
Pathology records of brain tumors resected at Department of Neurosurgery, Keio University Hospital, from 1990 through 2016 were reviewed, and patients who fulfilled the following criteria were included: (1) age ≥ 18 years old, (2) the original institutional diagnosis of World Health Organization (WHO) grade II or III glioma (lower-grade glioma, LrGG) according to WHO criteria at each time [2,3,4,5], (3) no prior radiotherapy or chemotherapy, (4) sufficient tumor samples for molecular analyses, (5) written informed consent, or, for the deceased patients and the patients who underwent surgical intervention before December 2000, opt-out consent via website. To exclude misdiagnosis by sampling error, biopsy cases were excluded. Formalin-fixed paraffin-embedded (FFPE) tissue blocks were obtained from tissue bank of the neurosurgery department and clinical pathology department.
Clinical data including age at diagnosis, sex, extent of resection, original histopathological diagnosis, survival information, and the date of the last contact were obtained from patients’ records. The data were frozen on November 2020.
Histopathological re-assessment
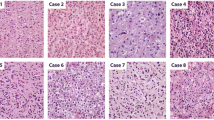
Because the diagnostic trends as well as criteria of histological classification of diffuse gliomas have changed over time, the histological classification of the tumors was re-assessed based on the morphological features by 2 neuropathologists (MS, KO) blinded to the original histological diagnoses as well as molecular features. Here, the re-assessment was based on the WHO2007 criteria [5], the most recent diagnostic criteria solely based on histology, because the aim of this study was to clarify the molecular and clinical characteristics of oligodendroglial morphology regardless of IDH-1p19q status. Special attention was paid to (1) the diagnosis of oligodendroglioma or anaplastic oligodendroglioma (pure oligodendroglial diagnosis) based on WHO2007, (2) the presence or absence of histological characteristics classic for oligodendroglioma (CFO) within a tumor: the presence of cellular monomorphism, round/ regular nuclei, presence of nodules, microcalcifications, microcysts and chicken wire vasculature [6] (Fig. 1). If a tumor shows such characteristics in part, for example, in at least one low-power field area, the tumor was judged to have CFO. A tumor for which diagnosis was oligoastrocytoma but has CFO feature in part was judged to have CFO despite the diagnosis of mixed oligoastrocytoma. Malignancy grade was also based on WHO2007. In case of discrepancy between the 2 neuropathologists, re-assessment was made until consensus has been met.
Molecular analyses
Deoxyribonucleic acid (DNA) extraction
Tumor DNA was extracted from microdissected pieces of FFPE sections. Microdissection was performed on consecutive sections based on hematoxylin–eosin (HE) staining and MIB-1 immunohistochemical staining (Monoclonal mouse anti-human Ki-67 Antigen Clone MIB-1, Dako, Glostrup, Denmark) to exclude intermixed non-neoplastic glial/vascular cells and hemorrhagic/necrotic regions [7].
Mutations in the IDH 1/2 genes
Tumor IDH mutation status was first assessed by immunohistochemistry (IHC) with an anti-IDH1R132H antibody (Mouse mAb Clone H09, Dianova, Hamburg, Germany) on FFPE sections. Tumors with negative result for the IHC were then analyzed for IDH1/2 mutations by Sanger sequencing as previously described [8].
Mutations in the ATRX gene
Mutations in the ATRX gene were assessed by immunohistochemistry. Immunohistochemistry for ATRX was performed with rabbit monoclonal anti-ATRX antibody (HPA001906, 1:200, Sigma-Aldrich, Missouri, United States) after antigen retrieval by microwave for 25 min in Tris–EDTA buffer (pH 9). In cases with equivocal staining, staining > 30% of nuclei was judged as positive for ATRX.
Mutations in the promoter region of the TERT gene
The 163 bp promoter region of the TERT gene flanking the known 2 mutation hot spots was amplified using the previously published primer pairs as previously described [9, 10]. Sanger sequence was performed bidirectionally.
Hypermethylation in the promoter region of the MGMT gene
The methylation status of the promoter region of the MGMT gene was assessed using methylation-specific PCR as previously described [7]. Bisulfate reaction and subsequent PCR were performed with the EZ DNA Methylation-Direct kit (Zymo Research Corp., Orange, California, USA) and the previously published primer pairs [7].
Comparative genomic hybridization
CNAs of the tumors were assessed by comparative genomic hybridization (CGH) as previously described [11, 12]. Briefly, DNA was extracted from FFPE sections by microdissection of truly neoplastic tissue referring to the HE staining and MIB-1 immunohistochemistry of the consecutive sections. The extracted DNA was amplified using degenerate oligonucleotide primed-polymerase chain reaction (DOP-PCR) to provide wider template. Tumor DNA was labeled with digoxigenin (DIG)-11-dUTP (Roche, Mannheim, Germany), and the reference DNA was amplified from 50 ng of normal male or female DNA and labeled with biotin-dUTP (Roche). The probe mixture was hybridized to the normal metaphase spread (Vysis, Downer Grove, Illinois, USA). The spread was then hybridized with fluorescein isothiocyanate-conjugated anti-DIG-antibody (Roche) and rhodamine-conjugated avidin (Roche). The ratios of fluorescence intensity along the chromosomes were analyzed by Cytovision analysis system (Applied Imaging, San Jose, California, USA).
Next-generation sequencing-based gene panel tests
Next-generation sequencing (NGS)-based cancer gene panel test was performed with PleSSision panel in IDHmut/codel LrGGs. PleSSision panel is composed of 160 genes known to be relevant with cancer development [13, 14]. Tumor DNA was extracted from FFPE block containing minimum of 20 ng DNA.
DNA methylation-based classification of the IDH-wildtype LrGGs
Classification based on the DNA methylation profiling is a new diagnostic tool that provide robust and accurate diagnosis of CNS tumors. We performed methylome analysis using Infinium Methylation EPIC BeadChip Kit (Illumina, San Diego, CA) using the protocol previously described to characterize IDH-wildtype LrGGs [15, 16], which are supposed to include various tumor types such as circumscribed gliomas and diffuse gliomas with molecular signatures of glioblastoma [17]. Methylation data was analyzed using specific algorithm for central nervous system tumors that was developed by German Cancer Research Center (DKFZ) (https://www.molecularneuropathology.org/mnp) [18].
Statistical analysis
Overall survival (OS) was calculated from the beginning of treatment, i.e., from the date of the initial surgery. Cases with loss to follow-up were censored at the last day of follow-up. OS was analyzed and compared using Kaplan–Meier method and log-rank test. Significance levels for all tests were two sided and were set at 0.05. For the factors with a p value ≤ 0.1 in univariate analyses, the effect on survival was also estimated by multivariate analysis using Cox proportional hazard model and MANOVA test. The association between oligodendroglial morphology and MGMT promoter status was assessed by Chi-square test. All statistical analyses were performed by SPSS 26 (IBM, New York, United States).
Results
Oligodendroglial morphology correlates with 1p19q codeletion
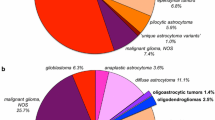
Ninty-three consecutive patients who fulfilled the eligibility criteria with diffuse LrGGs were included in the study. There were 51 men and 42 women, and the median age at initial surgery was 38 years (15–84). The histological diagnoses after the re-assessment based on WHO2007 were diffuse astrocytoma (DA) in 4 cases, anaplastic astrocytoma (AA) in 21 cases, oligodendroglioma (OD) in 14 cases, anaplastic oligodendroglioma (AO) in 15 cases, oligoastrocytoma (OA) in 10 cases, anaplastic oligoastrocytoma (AOA) in 29 cases (Table 1). Thus, 29 tumors were diagnosed as pure oligodendroglial tumor (oligodendroglial histological diagnosis) based on WHO2007, and CFO was stringently judged by consensus of 2 neuropathologists in 20 tumors; 2 in oligodendroglioma, 12 in anaplastic oligodendroglioma, 2 in oligoastrocytoma, 4 in anaplastic oligoastrocytoma. On the other hand, 93 tumors were classified into IDHmut/codel in 27, IDHmut/noncodel in 47, and IDHwild in 19 by definition of WHO2016. The detailed comparison between the original institutional diagnoses, re-assessed diagnoses based on WHO2007, and the diagnoses based on WHO2016 will be reported elsewhere.
1p/19q codeletion was present in 16 of the 29 tumors (55%) with oligodendroglial histological diagnoses, i.e., oligodendrogliomas or anaplastic oligodendrogliomas, 11 of 39 cases of those with mixed diagnoses (28%), and none of those with astrocytic diagnoses. On the other hand, 1p/19q codeletion was present in 13 of the 20 tumors with CFO (65%), and 14 of the 73 tumors without CFO (19%). There was significant association between 1p19q codeletion status with oligodendroglial histological diagnoses based on WHO 2007 (p < 0.0001), as well as between 1p19q codeletion and presence of CFO (p < 0.0001).
Median follow-up of the alive patients was 4.7 years. As with the literature, the IDH-1p19q-based grouping of LrGGs significantly correlated with OS, with IDHmut/codel tumors being most favorable (median OS: IDHmut/Codel 14.4 years, IDHmut/Noncodel 12.8 years, IDHwild 2.9 years, IDHmut/codel vs IDHmut/noncodel; p = 0.039, IDHmut/codel vs IDHwild p < 0.0001; IDHmut/noncodel vs IDHwild p = 0.004). On the other hand, neither the oligodendroglial histological diagnoses nor CFO was correlated with patient OS in the entire cohort; median OS of the tumors with oligodendroglial histological diagnoses being 14.4 years as compared with 12.8 years in those with mixed or astrocytic histology (p = 0.217), and 14.4 years in tumors with CFO versus 13.6 years without CFO (p = 0.368).
The IDH-1p19q-based grouping was correlated with the methylation status of the MGMT gene, with methylated promoter in 23 of 27 IDHmut/codel, in 28 of 47 IDHmut/noncodel, and in 6 of 19 IDHwild tumors (p = 0.001). On the other hand, the oligodendroglial histological diagnosis, but not CFO, was marginally associated with MGMT promoter methylation (p = 0.051, p = 0.155). However, neither oligodendroglial histological diagnosis nor presence of CFO was independently associated with MGMT promoter methylation by multivariate analysis with 1p/19q codeletion (p = 0.655, 0.851). To further examine the significance of oligodendroglial morphology itself, the subsequent analyses were performed within each of the subgroup based on IDH-1p19q status.
Prognositc relevance of oligodendroglial morphology within each of IDH-1p19q subgroup
In the 27 IDHmut/codel LrGGs, 16 (grade II: 7, grade III: 9) were diagnosed as either oligodendroglioma or anaplastic oligodendroglioma (oligodendroglial histological diagnosis, 59%) by definition of WHO2007, and 13 (grade II: 3, grade III: 10) were judged to have CFO features (48%). Patients with the tumors with oligodendroglial histological diagnosis had significantly longer OS than those with mixed oligoastrocytomas in the patients with IDHmut/codel tumors (p = 0.023) (Fig. 2A). Although the trends were also shown, the association was not statistically significant after adjustment for histological grade or patient age (Table 2). On the other hand, presence of CFO was not associated with OS in the patients with IDHmut/codel tumors (p = 0.594) (Fig. 2B). There were no correlations between MGMT promoter methylation status and oligodendroglial histological diagnosis or presence of CFO within the IDHmut/codel subgroup (p = 0.936, 0.125).
A Kaplan–Meier analysis of overall survival (OS) according to oligodendroglial diagnosis by WHO2007 in IDHmut/codel group. Green line denotes patients with pure oligodendroglial diagnosis, and blue line those with mixed oligoastrocytic diagnosis. The oligodendroglial diagnosis was associated with longer OS as compared with mixed oligoastrocytic tumors (p = 0.023). B Kaplan–Meier analysis of overall survival (OS) according to presence of CFO features in IDHmut/codel group. Green line denotes patients with tumors with CFO features, and blue line those without. There was no significant difference in OS between tumors with CFO features and those without it (p = 0.594)
In the 47 IDHmut/noncodel LrGGs, 11 were diagnosed as either oligodendroglioma or anaplastic oligodendroglioma (23%), and 5 were judged to have CFO (11%). Neither the oligodendroglial histological diagnoses nor presence of CFO was significantly associated with OS within the IDHmut/noncodel LrGGs by Kaplan–Meier and log-rank analyses (p = 0.177, 0.618, respectively) (Fig. 3A, B). The lack of association was also shown by Cox analyses, and the hazards for death were even higher with oligodendroglial morphology (Table 2). Moreover, neither oligodendroglial histological diagnosis nor presence of CFO was significantly associated with MGMT promotor methylation status in this subgroup (p = 0.086, 0.325, respectively).
A Kaplan–Meier analysis of overall survival (OS) according to oligodendroglial diagnosis by WHO2007 in IDHmut/noncodel group. Green line denotes patients with pure oligodendroglial diagnosis and blue line denotes those with wither mixed oligoastrocytic or astrocytic diagnoses by WHO2007. Pure oligodendroglial diagnosis was not associated with longer OS (p = 0.177). B Kaplan–Meier analysis of overall survival (OS) according to presence of CFO features in IDHmut/noncodel group. Green line denotes patients with tumors with CFO features, and blue line denotes those without it. Presence of CFO was not associated with longer OS (p = 0.618)
In the 19 IDHwild LrGGs, 2 were diagnosed as either oligodendroglioma or anaplastic oligodendroglioma (11%), and 2 were judged to have CFO (11%) (one case with both: anaplastic oligodendroglioma and CFO). Neither the oligodendroglial histological diagnoses nor presence of CFO was associated with OS (p = 0.123, 0.726) or MGMT promoter methylation (both p = 0.310) within the IDHwild LrGGs.
Molecular background of oligodendroglial morphology
In total of 93 tumors, ATRX mutations as assessed by immunohistochemistry were noted in 39 tumors; none of 27 IDHmut/codel, 34 of 47 IDHmut/noncodel, and 5 of 19 IDHwild tumors. TERT promoter mutations were noted in all of 27 IDHmut/codel, none of 47 IDHmut/noncodel, and 9 in 19 IDHwild tumors. Lack of ATRX mutations was not associated with either oligodendroglial histological diagnosis or presence of CFO in IDHmut/noncodel tumors (p = 0.132, 0.514).
CNAs detected in more than 10% of 93 tumors except for 1p19q codeletion included gain of 7q31.3-ter (n = 15), gain of 19pter-13.3 (n = 12), and gain of 8q22-24 (n = 10). Of these, none was significantly associated with either of oligodendroglial histological diagnosis or CFO. CNAs found in ≥ 10% of the 27 IDHmut/codel LrGGs were loss of 4pter-15.1 (n = 6), loss of 18q (n = 7), gain of 7 (n = 5), gain of 11q23.1-ter (n = 5) and loss of 18p (n = 4). Of these, none was significantly associated with oligodendroglial histological diagnosis or presence of CFO in IDHmut/codel tumors. The mean number of CNAs except 1p/19q codeletion in the IDHmut/codel tumors with oligodendroglial histological diagnosis was 2.7 (0–10) and that of the tumors with mixed histology was 1.7 (0–6). Therefore, there was no significant difference in the number of CNAs between pure oligodendroglial tumors and the tumors of mixed diagnosis (p = 0.99). Similarly, there was no significant difference in the number of CNAs between IDHmut/codel tumors with CFO and those without (mean 2.6 with CFO vs 2.0 without CFO, p = 0.775).
CNAs found in ≥ 10% of the 47 IDHmut/noncodel LrGGs were gain of 7q31.3-ter (n = 15), gain of 8q22-24 (n = 10), gain of 19pter-13.3 (n = 7), gain of 1pter-35 (n = 6), gain of 10pter-12 (n = 6), loss of 9p24 (n = 6) and loss Xp21-13 (n = 5). Of these, gain of 8q was significantly associated with oligodendroglial histological diagnosis (p = 0.049), and gain of 1p and gain of 7q were numerically more frequent in the tumors with oligodendroglial histological diagnosis (p = 0.099, 0.09). However, none of the CNAs was significantly associated with presence of CFO.
Prognostic relevance of molecular factors within each of IDH-1p19q subgroup
Among the CNAs found in ≥ 10% of the IDHmut/codel LrGGs, none was significantly associated with OS by Kaplan–Meier estimation and log-rank analysis. Similarly, neither ATRX mutations nor any of the CNAs were associated with OS in IDHmut/noncodel LrGGs (ATRX p = 0.530).
Neither ATRX mutations nor TERT promoter mutations were associated with patient OS in IDHwild LrGGs (ATRX p = 0.280, TERT p = 0.980) (Supplemental Fig. 1A, B). Of these, 8 cases showed no CAN; however, lack of CNA was not associated with OS (p = 0.471). Among the CNAs found in ≥ 10% of the IDHwild LrGGs, loss of 4q32-ter (p = 0.002), gain of 7qcen-11.2 (p = 0.004), gain of 7q31.3-ter (p = 0.004), gain of 8qcen-12 (p = 0.004), loss of 13q21.3 (p = 0.002), and gain of 17q25 (p = 0.004) were associated with OS by Kaplan–Meier estimation and log-rank analysis.
NGS-based gene panel test for IDHmut/codel LrGGs
The finding that morphological diagnosis of oligodendroglioma might be favorably associated with OS within the IDHmut/codel tumors facilitated us to investigate if any association between oligodendroglial morphology and certain gene mutations in these tumors. PleSSision cancer gene panel test was successfully performed in 20 of the 27 IDHmut/codel LrGGs. The panel test was not feasible in the 7 tumors due to low DNA quality or the unavailability of the extra tumor specimens. In all, mutations were found in the 15 genes, and, among those, 5 genes were found mutated repetitively; CIC in 6, FUBP1 in 3, NOTCH1 in 3, PBRM in 2 cases, and NF1 in 2 cases. However, none of mutation status was associated with oligodendroglial histological diagnosis by definition of WHO2007. On the other hand, mutations in the FUBP1 gene were associated with poor OS in patients with IDHmut/codel LrGGs (p = 0.005), and mutations in CIC showed trends towards poor OS (p = 0.061) (Supplemental Fig. 2A, B).
DNA methylation-based classification of IDH-wildtype LrGGs
The IDHwild LrGGs are suggested to be composed of heterogeneous tumor types including tumors with molecular features of glioblastomas and circumscribed tumors such as pleomorphic xanthoastrocytoma and glioneuronal tumors. To explore the accurate diagnosis, IDH-wildtype tumors for which extra DNA was available were classified using Infinium MethylationEPIC BeadChip Kit and the Classifier provided by DKFZ (https://www.molecularneuropathology.org/mnp). Unfortunately, the reliable results were obtained only in 3 tumors possibly due to small amount of available DNA and/or degradation of DNA in old specimens. The methylation-based classification of the 3 tumors included (anaplastic) pleomorphic xanthoastrocytoma and ganglioglioma; however, the profile of the other one did not fit into any of the known methylation class (Supplemental Fig. 3).
Discussion
In the present study, 93 LrGGs treated in a single institute were re-assessed for their morphology-based diagnosis, and the clinical significance and molecular background of oligodendroglial morphology were investigated within each of the IDH-1p/19q-based 3 molecular group as well as in total 93 cases. As a result, oligodendroglial morphology, as assessed by histological diagnosis based on WHO2007 and presence of CFO, was correlated with 1p/19q codeletion expectedly in the entire cohort. As to the association between oligodendroglial morphology and patient prognosis, the oligodendroglial histological diagnosis was not independently associated with patient OS in any molecular subgroups, but with a signal of possible favorable association in univariate analysis in IDHmut/codel tumors. Unfortunately, neither oligodendroglial diagnosis or presence of CFO was predictive of methylation status of the MGMT gene in any of the 3 molecular subgroups. Interestingly, gain of 8q was significantly associated with oligodendroglial diagnosis (oligodendroglioma or anaplastic oligodendroglioma based on WHO2007) in IDHmut/noncodel tumors.
Although the literature suggests that IDH-1p/19q status is more relevant than histological phenotype to the prognoses of the patients with LrGGs [20, 21], the oligodendroglial morphology itself might be prognostic for patients with LrGGs [6, 22]. For example, Gianinni et al. reported that, in the cohort of phase III clinical trial for grade III gliomas, the classic oligodendroglial morphology was associated with prolonged OS in the multivariate analysis with 1p/19q status in the entire cohort [6]. In the present study, by the analyses in each of the 3 molecular subgroups, either the oligodendroglial histological diagnosis or presence of CFO was not associated with patient OS in IDHmut/noncodel tumors. On the other hand, though not statistically significant in the multivariate analyses, there was a trend of favorable association between the oligodendroglial histological diagnosis and patient OS in IDHmut/codel tumors. Indeed, in the previous study, OS was numerically longer in CFO patients than non-CFO in 1p/19q codeleted grade III gliomas, but without statistical significance. Thus, considering the subjective nature of morphology-based diagnosis, the results of the present study and the previous study did not contradict, and the significance of the oligodendroglial phenotype might be different according to molecular subgroup. Altogether, the oligodendroglial morphology might be nothing more than a surrogate marker for 1p/19q codeletion in LrGGs, leaving the possibility of marginal favorable association with patient OS only in IDHmut/codel tumors.
The NGS panel analyses of the IDHmut/codel tumors showed that the CIC and FUBP1 mutations might be correlated with poor patients’ prognoses, however, a previous study suggested the contrary association [23, 24]. On the other hand, although the NOTCH1 mutations was not associated with patients’ OS in the present study, a previous study with 141 and 139 IDHmut/codel tumors from Japan and The Cancer Genome Atlas (TCGA) data, respectively, suggested the correlation of the NOTCH1 mutations and unfavorable survival in Japanese data but not in TCGA [25]. Our results in terms of the relevance of mutations of those genes might not be conclusive due to the small number of cases, and the reasons for the discrepancy are not clear. Further investigation in the cohort with large number of cases and uniform treatment is warranted.
Interestingly, the present study showed that gain of 8q was significantly associated with oligodendroglial diagnosis in IDHmut/noncodel tumors. On the other hand, a previous study reported that loss of 19q was associated with oligodendroglial morphology as well as favorable prognosis in IDH-mutant astrocytomas [26]. In the present study, there was only one case with 19q loss in IDH mut/noncodel LrGG which was diagnosed as anaplastic oligoastrocytoma by histological re-assessment.
In 19 IDH-wildtype LrGGs, only 3 cases were diagnosed to have pure oligodendroglial diagnosis and/or CFO for each. Neither oligodendroglial diagnosis or presence of CFO was associated with patients’ prognoses or any of the molecular factors. Therefore, the oligodendroglial histological features are not prognostic in the IDH-wildtype LrGGs. Also, none of the molecular factors was independently associated with pure oligodendroglial diagnosis or presence of CFO. Unfortunately, classification based on DNA methylation array was successful in only 3 IDH-wildtype LrGGs; (anaplastic) pleomorphic xanthoastrocytoma, ganglioglioma, and the one which apparently not fit into any of the methylation class. Those findings substantiate that IDH-wildtype LrGGs are composed of heterogeneous tumor types including the ones not established yet so far. Although loss of 4q32-ter, gain of 7qcen-11.2, gain of 7q31.3-ter, gain of 8qcen-12, loss of 13q21.3, and gain of 17q25 were associated with OS in IDHwild LrGGs, given the heterogenous nature of this subgroup, further investigation is warranted.
The vast majority of IDHmut/codel, the majority of IDHmut/noncodel, and about 40% in IDH-wildtype LrGGs were known to have methylated promoter in the MGMT gene [27]. However, unfortunately, the present study demonstrated that neither oligodendroglial diagnosis nor presence of CFO was predictive of methylation status of the MGMT gene in any of the 3 molecular subgroups.
Limitations of the study
We regret that the number of tumors included in the present study might be small to draw definite conclusion for some queries including the relevance of the CIC, FUBP1, and NOTCH1 mutations to patients’ prognoses. Moreover, because of the retrospective nature of the study, treatment for the cases was not uniform, which could affect the results with survival outcome. However, the study was performed in single institutional consecutive cohort, and the difference in treatment between cases was minimal. Moreover, OS, not PFS, is the robust survival outcome following all of the available treatments (both radiotherapy and chemotherapy must have been prescribed in all of the deceased cases), and the present study showed, for the first time, that the oligodendroglial histological phenotype is not associated with patient survival in IDHmut/noncodel tumors. On the other hand, because of the above-mentioned limitations, further studies are warranted for the relevance of the oligodendroglial phenotype to patient survival in IDHmut/codel tumors.
Conclusion
The present study suggested that oligodendroglial morphology, as assessed by histological diagnosis based on WHO2007 and presence of CFO, was not independently associated with either patient OS or methylation status of the MGMT gene in LrGGs, leaving the possibility of marginal favorable association only in IDHmut/codel tumors. These findings might suggest that oligodendroglial morphology is nothing more than a surrogate marker for 1p/19q codeletion, and substantiate the importance of the molecular classification in LrGGs.
Abbreviations
- ATRX:
-
Alpha-thalassemia
- CFO:
-
Classic for oligodendroglioma
- CIC:
-
Capicua transcriptional repressor
- CGH:
-
Comparative genomic hybridization
- FUBP1:
-
Far upstream element-binding protein 1
- IDH:
-
Isocitrate dehydrogenase
- IDHmut/codel:
-
IDH-mutant and 1p/19q codeleted
- IDHmut/noncodel:
-
IDH-mutant and 1p/19q noncodeleted
- IDHwild:
-
IDH-wildtype
- IHC:
-
Immunohistochemistry
- LrGG:
-
Lower-grade glioma, i.e., WHO grade II or III glioma
- MGMT:
-
O6-methylguanine-DNA methyltransferase
- NGS:
-
Next-generation sequencing
- OS:
-
Overall survival
- PCR:
-
Polymerase chain reaction
- TERT:
-
Telomere reverse transcriptase
- WHO:
-
World Health Organization
- WHO2007:
-
WHO classification of tumors of the central nervous system 4th edition
- WHO2016:
-
WHO classification of tumors of the central nervous system, revised 4th edition
References
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2016) WHO classification of tumours of the central nervous system. International Agency for Research on Cancer, Lyon
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2007) WHO classification of tumours of the central nervous system. International Agency for Research on Cancer, Lyon
Zulch KJ (1979) Histological typing of tumours of the central nervous system. Switzerland, Geneva
Kleihues P, Burger P, Scheithauer BW (1993) Histological typing of tumours of the central nervous system. Springer, Berlin
Kleihues P, Cavenee W (2000) Pathology and genetics of tumours of the nervous system. IARC Press, Lyon
Giannini C, Burger P, Berkey BA et al (2008) Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol 18(3):360–369
Ezaki T, Sasaki H, Hirose Y, Miwa T, Yoshida K, Kawase T (2011) Molecular characteristics of pediatric non-ependymal, non-pilocytic gliomas associated with resistance to temozolomide. Mol Med Rep 4(6):1101–1105
Hayashi S, Sasaki H, Kimura T et al (2015) Molecular-genetic and clinical characteristics of gliomas with astrocytic appearance and total 1p19q loss in a single institutional consecutive cohort. Oncotarget 6(18):15871–15881
Koelsche C, Wohrer A, Jeibmann A et al (2013) Mutant BRAF V600E protein in ganglioglioma is predominantly expressed by neuronal tumor cells. Acta Neuropathol 125(6):891–900
Nakagawa Y, Sasaki H, Ohara K et al (2017) Clinical and molecular prognostic factors for long-term survival of patients with glioblastomas in single-institutional consecutive cohort. World Neurosurg 106:165–173
Hirose Y, Aldape K, Takahashi M, Berger MS, Feuerstein BG (2001) Tissue microdissection and degenerate oligonucleotide primed-polymerase chain reaction (DOP-PCR) is an effective method to analyze genetic aberrations in invasive tumors. J Mol Diagn 3(2):62–67
Miwa T, Hirose Y, Sasaki H, Ezaki T, Yoshida K, Kawase T (2011) Single-copy gain of chromosome 1q is a negative prognostic marker in pediatric nonependymal, nonpilocytic gliomas. Neurosurgery 68(1):206–212
Saotome K, Chiyoda T, Aimono E et al (2020) Clinical implications of next-generation sequencing-based panel tests for malignant ovarian tumors. Cancer Med 9(20):7407–7417
Aimono E, Nishihara H (2020) Required human and system resources for genomic medicine. Yakugaku Zasshi 140(5):651–655
Makabe T, Arai E, Hirano T et al (2019) Genome-wide DNA methylation profile of early-onset endometrial cancer: its correlation with genetic aberrations and comparison with late-onset endometrial cancer. Carcinogenesis 40(5):611–623
Ohara K, Arai E, Takahashi Y et al (2018) Feasibility of methylome analysis using small amounts of genomic DNA from formalin-fixed paraffin-embedded tissue. Pathol Int 68(11):633–635
Brat DJ, Aldape K, Colman H et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.” Acta Neuropathol 136(5):805–810
Capper D, Jones DTW, Sill M et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555(7697):469–474
Brat DJ, Aldape K, Colman H et al (2020) cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139(3):603–608
Suzuki H, Aoki K, Chiba K et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47(5):458–468
Brat DJ, Verhaak RG, Cancer Genome Atlas Research Network et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372(26):2481–2498
McDonald JM, See SJ, Tremont IW et al (2005) The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer 104(7):1468–1477
Cahill DP, Louis DN, Cairncross JG (2015) Molecular background of oligodendroglioma: 1p/19q, IDH, TERT, CIC and FUBP1. CNS Oncol 4(5):287–294
Morin GB, Chan JA, Cairncross JG, Marra MA (2012) Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 226(1):7–16
Aoki K, Nakamura H, Suzuki H et al (2018) Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol 20(1):66–77
Otani R, Uzuka T, Higuchi F et al (2018) IDH-mutated astrocytomas with 19q-loss constitute a subgroup that confers better prognosis. Cancer Sci 109(7):2327–2335
van den Bent MJ, Dubbink HJ, Marie Y et al (2010) IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European organization for research and treatment of cancer brain tumor group. Clin Cancer Res 16(5):1597–1604
Acknowledgements
The present study was supported by Grant-in-Aid for Scientific Research (KAKENHI) by the Ministry of Education, Culture, Sports, Science and Technology (Grant no. 19104130). We greatly thank Ms. Naoko Tsuzaki from the Department of Neurosurgery at Keio University School of Medicine for her technical assistance. We thank Dr. Masahiro Toda from the Department of Neurosurgery at Keio University School of Medicine for general supervision.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pareira, E.S., Shibuya, M., Ohara, K. et al. The oligodendroglial histological features are not independently predictive of patient prognosis in lower-grade gliomas. Brain Tumor Pathol 39, 79–87 (2022). https://doi.org/10.1007/s10014-022-00426-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-022-00426-5