Abstract
Bacillus thermocatenulatus lipase (BTL2), a member of the isolated lipase family known as thermoalkalophilic lipases, carries potential for industrial applications owing to its ability to catalyze versatile reactions under extreme conditions. This study investigates the molecular effects of distinct solvents on the stability of BTL2 at different temperatures, aiming to contribute to lipase use in industrial applications. Initially, molecular dynamic (MD) simulations were carried out to address for the molecular impacts of distinct solvents on the structural stability of BTL2 at different temperatures. Two lipase conformations representing the active and inactive forms were simulated in 5 solvents including water, ethanol, methanol, cyclohexane, and toluene. Low temperature simulations showed that polar solvents led to enhanced lid fluctuations compared with non-polar solvents reflecting a more dynamic equilibrium between active and inactive lipase conformations in polar solvents including water, while the overall structure of the lipase in both forms became more rigid in non-polar solvents than they were in polar solvent. Notably, the native lipase fold was maintained in non-polar solvents even at high temperatures, indicating an enhancement of lipase’s thermostability in non-polar organic solvents. Next, we conducted experiments for which BTL2 was expressed in a heterologous host and purified to homogeneity, and its thermostability in different solvents was assessed. Parallel to the computational findings, experimental results suggested that non-polar organic solvents contributed to BTL2’s thermostability at concentrations as high as 70% (v/v). Altogether, this study provides beneficial insights to the lipase use under extreme conditions.

Graphical Abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) are group of enzymes acting on carboxylic ester bonds, by either breaking or forming it. They catalyze a wide range of chemical reactions including hydrolysis and synthesis of esters and transesterification reactions [1,2,3,4]. Owing to the versatility of the reactions that can be catalyzed by lipases, they carry paramount potential for industrial applications such as oleochemistry, biofuel, and pharmaceutical production [5,6,7,8].
Notwithstanding such catalytic advantages offered by lipases, harsh conditions encountered in industrial processes, particularly elevated temperatures and high exposure to organic solvents, may limit their industrial use. Basically, due to their biochemical nature, proteins are generally unstable at high temperatures and in the presence of organic solvents and denature under such extreme conditions [9]. Nevertheless, this concept holds for most of the proteins; studies over the last decades have led to a shift in this paradigm of protein instability at high temperatures and in organic solvents [10, 11]. As such, some of the enzymes including lipases are shown to become thermo-stabilized in non-aqueous solvents. For example, the porcine pancreatic lipase was reported to have a half-life of several hours at 100 °C and in organic solvents whereas it underwent thermal denaturation only within few seconds in aqueous medium [12].
Designing lipase variants with optimal stabilities for industrial processes requires a complete understanding of the molecular mechanism behind thermal and organic solvent stability. Although numerous studies have investigated selectivity and stability of thermoalkalophilic lipases, [13,14,15,16,17] thermostability of these lipases has not been studied in different solvents. Particularly, a molecular level understanding of lipases’ thermostability in different organic solvents would contribute to the design of highly stable thermoalkalophilic lipase variants at elevated temperatures and in organic solvents.
In this study, we combined both theoretical and experimental approaches to understand how different organic solvents affect the structure and activity of the lipase originating from the microbial species Bacillus thermocatenulatus (BTL2). BTL2 which was selected as a representative for the thermoalkalophilic lipase family is a viable candidate for prospective industrial applications owing to its ability to catalyze versatile reactions [18]. For the theoretical studies, molecular dynamic (MD) simulations of both of the lipase conformations representing the active/open (PDB ID: 2 W22) [19] and the inactive/closed form (PDB ID: 1KU0) [20] of thermoalkalophilic lipases were carried out. Both of the conformations were solvated in 5 different solvents and simulated at 2 different temperatures. Overall, a total of 20 different systems were generated and analyzed by MD simulations which provided us with the significant observation of enhanced thermostability in non-polar solvents. Overall, comparing the experimental results with the insights gained from simulations provided us with a better understanding of the behavior of lipases in different organic solvents. The results obtained by this study shed light on the organic solvent stability of thermoalkalophilic lipases at elevated temperatures reflecting the importance of computational methods for understanding protein stability and activity at extreme conditions.
Material and methods
Structure preparation
The active/open (PDB ID: 2 W22, 2.2 Å, R-factor = 0.182) and inactive/closed (PDB ID: 1KU0, 2.0 Å, R-factor = 0.185) conformations of thermoalkalophilic lipases were retrieved from the PDB. Prior to simulations, the water molecules were removed from the structure, whereas the metal ions, Zn+2, and Ca+2 were kept. The co-factors were also removed from the structures prior to simulations. Among two molecules, EGC acts as a substrate analog which led capturing of the open conformation. The reason of removing EGC from the structures is to test the movement of the open conformation only in response to different solvents. None of the structures contain atoms with multiple occupancies. For the dimer closed structure, chain A was selected for the simulations because no apparent discrepancies were noted between two subunits. The open and closed conformations were fully solvated with water, ethanol, methanol, toluene, or cyclohexane. The systems were generated by placing the lipase at the center of the box which was then filled with the solvent molecules. Given the large size of some of the solvent molecules, they were initially placed at least 2 Å apart from each other to allow translational and rotational movements during equilibration [21].The solvation box dimensions were approximately 45 Å in each direction (x, y, z), and the protein was located at least 10 Å from the box edges. All of the solvated systems were neutralized by Na+ and Cl− ions.
Molecular dynamic simulations
The resulting systems of open and closed lipase structures in 5 different solvents were fully energy-minimized by the default minimizer of the NAMD algorithm which uses conjugate gradient and line search algorithms and equilibrated by initially NVT and then NPT ensembles [22]. A minimization of 40,000 steps followed by 0.25 ns of equilibration (NVT) of solvent molecules was performed for all of the systems wherein the protein atoms were fixed. Next, the same minimization and equilibration steps were performed without fixing the protein. Then, an equilibration simulation (NPT) was performed for all of the systems until a stable RMSD pattern was monitored for protein backbone.
All the simulations were carried out by the CHARMM36 all atom force field with map correction, [23] with a time-step of 2 fs and periodic boundary conditions in all dimensions [24, 25]. For water, TIP3P model was used [26]. The simulations were performed for 100 ns at 310 K and at 450 K using Langevin piston pressure method and Langevin dynamics temperature [27]. The particle-mesh Ewald (PME) method was used in the calculation of the electrostatic interactions with 12 Å non-bonded cutoff [28].
Data analysis
For visualization of the structures and the trajectories, VMD is used [29]. The trajectories were analyzed by means of backbone RMSD, carbon alpha (Cα) fluctuations, atom distances, and dihedral angles for which plugins of Visual Molecular Dynamic (VMD) program were used. Analyses of Essential Dynamics (ED) and Dynamic Cross Correlation Map (DCCM) were performed on the Cα atoms by the Bio3D package of R [30].
Expression and purification of BTL2 lipase
As a representative of the thermoalkalophilic lipase family, the lipase originating from B. thermocatenulatus lipase (BTL2) was chosen. Escherichia coli BL21 (DE3) cells were transformed with the pMCSG7 plasmid harboring the 1161 bp fragment encoding the mature BTL2 lipase gene [31]. The cells were grown overnight on LB Agar plates containing 100 μg/ml ampicillin at 37 °C. Then, a single colony was cultivated at 37 °C in LB starter culture containing 100 μg/ml ampicillin. For expression of the native lipases, an auto-induction protocol was applied with slight modifications to the Studier et al. [32]. Expression was carried out at 30 °C with vigorous shaking (165 rpm) in the Studier media of pH 7.0 which supplemented with 100 μg/ml of ampicillin. After 18 h of incubation with shaking, the cells were harvested by centrifugation at 5800 rpm for 20 min and 4 °C. Cell pellets were re-suspended in 20-mM sodium phosphate buffer at pH 7.4, and the suspension was incubated at room temperature for 30 min with gentle shaking. The cells were lyzed mechanically by sonication on ice for 6 min with the pulse on for 15 s and off for 45 s. In order to remove the cell debris, the crude cell free extract was centrifuged at 10,000 rpm for 45 min at 4 °C. For purification, the clarified extract was loaded onto a 2-ml Nickel coated HisTrap™ column (Amersham Biosciences, UK). A 20-mM sodium phosphate buffer containing 50 or 500 mM imidazole was used for binding or elution of the poly histidine (6xHis) tagged protein, respectively. The elution fractions were loaded onto a PD-10 desalting column (Amersham Biosciences, UK) for imidazole removal and buffer-exchange with sodium phosphate (50 mM, pH 7.5). The protein solution was concentrated to a final concentration of 2.50 mg/ml. The amount of protein was quantified by Bradford assay [33]. The expression and purification of lipase (BTL2) were confirmed by 12% SDS-PAGE gel.
Lipase assays
Enzyme activity was measured in a 96-well black micro-titer plate using 6 nM of lipase and the fluorescence substrate of 4-methylumbelliferyl butyrate. The reaction buffer contained 25 mM of sodium phosphate at pH 7.00. The substrate fluorescence was measured by the excitation wavelength of 355 nm and an emission wavelength of 460 nm in a kinetic fashion for every 30 s of an hour. The measurements were performed in triplicates and the initial velocities are calculated by using the software of the fluorimeter. A 600 nM of freshly prepared BTL2 were incubated in six different organic solvents (ethanol, methanol, acetone, 2-propanol, cyclohexane, and toluene) at different concentrations (0–90% v/v) for 30 min at 25 °C and in water, ethanol, and cyclohexane at different concentrations (25–70% v/v) for 30 min at 60 °C. The residual lipase activity of the one-hundredth of the incubated enzyme solution (6 nM) was analyzed by enzyme assays in a similar manner.
Results and discussion
MD simulations for probing molecular impacts of organic solvents and high temperatures
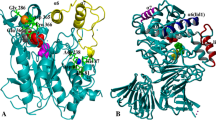
Lipases maintain an equilibrium between two distinct conformations. In aqueous environment, the lid domain hinders the active site of the lipases, rendering them inactive. When it comes in physical contact with an oil droplet, the lid opens at the water lipid interfaces by adsorption of the lid domain onto the lipid interface. This process known as interfacial activation is basically regulated by the structural rearrangements of the lid domain [34]. Although the size and composition of the lid domains differ among lipases, thermoalkalophilic lipases which comprise an isolated lipase family with high thermal and organic solvent stability possess a relatively large lid domain consisting of 2 α-helices (α6 and α7) spanning the amino acids from 169 to 239 in the closed structure (Fig. 1) [19]. Apparently, in this lipase family, lid opening requires an extensive structural movement for replacing 2 large helices covering the active site. In order to unravel the structural impact of solvents on thermoalkalophilic lipases, it would be essential to investigate both of the conformations.
Two different views of the aligned crystal structures of the open (2 W22) and the closed (1KU0) forms of thermoalkalophilic lipases. The lid portion was colored either red in the closed state or green in the open state. The Cα of the catalytic serine (S114) was also shown as an orange van der Waals sphere. Top panel shows the top view of the catalytic cleft and bottom panel shows the side view of the cleft
Owing to the high sequence conservation among the members of thermoalkalophilic lipases, all of the structures deposited in the protein databank (PDB) were investigated, and fortunately, both of the lipase structures at two distinct lid conformations were found to be resolved in separate crystallization studies (Fig.1). Bacillus stearothermophilus lipase (L1) were resolved in the closed state (PDB ID: 1KU0) while B. thermocatenulatus lipase (BTL2) were co-crystallized with substrate analogs in the open state (PDB ID: 2 W22) [19, 20]. Given the high sequence conservation between BTL2 and L1 (> 90%), these structures can be considered valid representations of the active and inactive conformations of thermoalkalophilic lipases respectively. Hence, these structures were recruited to MD simulations to study the impacts of solvents on structural stability. To specifically account for the solvent effects on thermostability, high temperature simulations were carried out at 450 K. Although the temperature of 450 K is an unrealistic experimental condition, such high temperature simulations are used to observe structural changes induced by elevated temperatures in relatively short time scales (ns). Examples of such “out of range” high temperature simulations are present as they offer a tradeoff between affordable computational sources and extreme conditions [35,36,37]. Overall, a total of 20 different simulations were carried out to address the molecular behavior of thermoalkalophilic lipases leading to organic solvent and thermal stability.
MD simulations revealed that the simulations showed a stabilized RMSD pattern for both of the open and closed state structures at 310 K, suggesting an equilibrium conformation for both states at 310 K (Fig. S1A). As anticipated, 450 K simulations showed an increased RMSD pattern fluctuating ~ 3 Å for the last 20 ns period of the simulations (Fig. S1B). Overall, MD simulations indicated equilibration for both of the lipase conformations that were solvated by 5 different solvents and simulated at 2 different temperatures. Hence, we underline the applicability of the molecular dynamic simulations carried out here for inspecting the molecular behavior of lipase in response to changes in the physical environment.
Polar solvents enhance lid fluctuations of both lipase conformations
The equilibrated closed structures showed enhanced fluctuations for the lid domain spanning the region between 169 and 239, particularly in the presence of polar solvents including water, ethanol, and methanol (Fig. 2a). Water led to the highest fluctuations which was followed by ethanol. Lastly, methanol was the polar solvent that led to a slight increase in the lid fluctuations. Fluctuations of the open conformation were strikingly very similar to the closed state such that the lid portion became more flexible in the polar solvents (Fig. 2a). On the other hand, the non-polar solvents including toluene, cyclohexane led to a much more rigid structure at 310 K regardless of the lid conformation (Fig. 2a).
Regardless the lid conformation, at 310 K, lipase showed significantly high SASA in the polar solvents while it remained tightly packed in the non-polar solvents (Fig. 3a). Concurrently, the radius of gyration analysis at 310 K showed that regardless the lid conformation, lipase maintains less compact structure in polar solvents than in non-polar ones (Fig. S2A). These observations propose that lipase structure regardless of its lid conformation becomes more flexible in the polar solvents while it stays intact in non-polar solvents.
The difference in protein flexibility between the polar and non-polar solvents is thermodynamically expected, since transferring a folded globular protein from water to less polar solvents like ethanol and methanol leads to burial of some of the protein polar groups resulting in an increase in the solvation free energy (ΔG) [38]. One the other hand, transferring a protein from a polar environment to nonpolar solvents such as cyclohexane and toluene leads to much higher solvation free energy (ΔG) [38]. This is mainly due to the burial of all the protein polar groups in order to overcome the unfavorable non-polar environment. This results in a very tightly packed and rigid structure of the protein in these nonpolar solvents [38]. Our observations on the change in the protein solvent accessible surface area (SASA) and radius of gyration (Rg) are in agreement with this solvation model (Figs.3, Fig. 4 and Fig S2A).
Non-polar organic solvents potentiate structural stability at high temperatures
Molecular dynamic simulations at 450 K were analyzed to evaluate thermal stability of lipase structure in different solvents. RMSF profiles at 450 K showed an increased mobility in polar solvents compared with the condition in non-polar solvents (Fig. 2b). Here we note a striking difference between the final structures obtained from 450 K simulations of polar and non-polar solvents. Regardless of the lid conformation, 450 K simulations led to a large structural destabilization i.e., thermal unfolding of the lipase structures in the polar solvents, while the structure was intact in non-polar solvents throughout 100 ns of simulation (Figs. 4 and S1B). Resistance to unfolding in the non-polar solvents was likely to be resulted from the reduced mobility in non-polar solvents (Fig. 2b). On the other hand, polar solvents such as water induced a thermal unfolding due to much increased mobility of the backbone (Figs. 2 and 4) The increased mobility of the lipase at high temperature in polar solvents can be easily spotted by the SASA analysis at 450 K (Fig. 3b). SASA of the lipase was increased to a higher level at which the compactness of the lipase structures is disturbed significantly (Fig. 2b). On the other hand, lipase remained tightly packed in the non-polar solvents despite this very high temperature. In line with these observations, the analysis of radius of gyration at 450 K showed that the two conformations of lipase maintain highly compact structure in non-polar solvents compared with polar solvents. The radius of gyration of lipase in polar solvents was high enough to indicate thermal unfolding (Fig S2B). Among the polar solvents, water had the lowest radius of gyration. This could be due to the unfolding of the protein to a collapsed or semi-aggregated structure as seen in Fig. 4c.
Flexibility disparities between the polar and non-polar solvents can also be seen by the structural visualizations representing the last frame of the 100-ns trajectory (Fig. 4). Polar solvents led to large alterations in the overall globular structure regardless of the lid conformation. Further Cα fluctuations reached to much higher values in the presence of polar solvents (Fig. 4c–e). Another notable point is that open conformation was more stable in ethanol than it was in water or methanol (Fig. 4c). This behavior was also seen in the RMSD profile of ethanol, which was similar to the polar solvents, particularly for the first period of the simulation (40 ns), while toward the end of simulation, it showed a similar behavior to the non-polar solvents (Fig. S1B). In ethanol, the lipase structures displayed some behavioral characteristics to both polar and non-polar solvents. Despite the polar nature of water, it did not produce similar results with ethanol. One of the possible reasons behind this observation would be the reactions induced in the presence of aqueous media such as Gln/Asn deamidation and peptide bond hydrolysis that are detrimental to enzyme stability [39].
The central of mass (COM) radial distribution function (RDF) analysis of the solvent molecules around the two lipase structures showed that the destabilizing effects of the polar solvents are beyond the protein surface. As seen in Figure S3A, at 310 K, regardless the lid conformation, the probability of polar solvents molecules being close to the protein core is higher than the non-polar solvent molecules. This could be due to the fact that polar solvent molecules are small enough to diffuse into the protein hydrophobic core disturbing its non-polar-buried groups. The diffusion rate of the polar solvent molecules increased at high temperature which could be due exposing of the protein hydrophobic core upon starting thermal unfolding (Fig. S3B).
Other observations supporting that non-polar organic solvents contribute to the structural stability of the lipase regardless of its conformation were obtained from the analyses of Essential Dynamics (ED) (Fig. S4) and Dynamic Cross Correlation Map (DCCM) (Fig. S5). ED analysis showed that the lipase spanned a limited conformational space when solvated in cyclohexane and toluene regardless of its lid conformation (Fig. S4). The lipase sampled a smaller number of conformations in these non-polar solvents at high temperature compared with the condition in polar solvents. DCCM analysis was also performed to assess the correlated motions in the simulations [40]. The lipase regardless of its lid conformation displayed similar positively and negatively correlated motions in non-polar solvent at both temperatures, underscoring higher thermostability in non-polar solvents. Overall, the ED analysis suggests higher stability of the lipase in non-polar solvents for both of the states. However, the correlated motions largely differed from low to high temperature simulations in the polar organic solvents, especially water and methanol. Notably, both ED and DCCM analyses suggested that the lipase structure in water was more tolerant to increased kinetic energy in its closed conformation compared with the open conformation. In contrast to this observation, the lipase is more thermo-tolerant in ethanol in its open form than its closed form. Given these theoretical analyses, one would expect an improved structural stability in non-polar organic solvents than in water, particularly at elevated temperatures. Overall, these results agree that the thermoalkalophilic lipase structure regardless of its lid conformation became stabilized in the presence of solvents of non-polar characteristics while they became destabilized in the presence of polar solvents with a notable exception in ethanol.
Polarity of the solvent mediates structural stability of lipase
One of the important interactions that play a critical role in interfacial activation is the formation of the salt bridge between Asp179 and Arg142 [19]. This salt bridge is located at the critical hinge region of the lid domain and stabilizes the open form by linking the lid domain to the hydrophobic core. Further two other salt bridges were found to be formed between Glu24:Lys208 and Arg93:Asp210. Although they are not specific to the active (open-lid) conformation, they are essential for the overall fold of the lipase. These salt bridges can be monitored to assess (i) the open-to-closed transition of the lid domain in which disruption of Asp179-Arg142 interaction would indicate lid closing, and (ii) the overall stability of the lipase.
The simulations performed at 310 K confirmed that all of 3 salt bridges were intact in all of the organic solvents throughout the entire simulation period, while they were slightly less persistent in water (Fig. 5a). Disrupted salt bridge that is characteristic to the open lipase structure indicates that the open lipase lid tends to adapt the closed conformation. Hence, parallel to the noted increase in the lid fluctuations at 310 K in polar solvents (Fig. 2a), we surmise that among the polar solvents water was the one that enabled the most dynamic equilibrium between open to closed state.
At 450 K, all of 3 interactions except Glu24:Lys208 were maintained in non-polar solvents while they were either completely or partially disrupted in the presence of polar solvents (Fig. 5b). Particularly, the salt bridge Glu24:Lys208 was completely disrupted in water while Arg93:Asp210 interaction was only partially affected in all of the polar solvent (Fig. 5b). Lastly, the salt bridge (Asp 179: Arg142) was lost in water and methanol while it was intact in ethanol and the non-polar solvents (Fig. 5b). As opposed to the observation in water at 310 K (Fig. 4a), the interaction between Glu24:Lys208 was largely separated in the presence of polar solvents, such that the distance reaching to values of 20–30 Å (Fig 5b). Here, we stress that such a large separation indicates thermal unfolding. This assertion was indeed supported by the RMSD measurements (Fig. S1B), SASA analysis (Fig. 3), and visual inspection of the lipase structure (Fig. 4c–e), all of which indicate thermal unfolding in the presence of polar organic solvents.
In addition to the salt bridges, we also monitored the catalytic machinery which is formed by a catalytic triad amino acids and an oxyanion hole [19]. The catalytic triad is formed by Ser114, Asp318, and His359 in BTL2 and Ser113, Asp317, and His358 in L1, and the oxyanion hole is formed by the amide groups of main chains of Phe17 and Gln115. In order to unravel the impact of organic solvents on the structural stability of the active site, we measured the radius of gyration of the catalytic machinery and distances between triad amino acids.
At 310 K, regardless the lid conformation, the distances between catalytic triad amino acids and the compactness of the catalytic machinery were found unaffected by the organic solvents (Fig. 6a and Fig. S6). We note that both of the catalytic triad distances and the radius of gyration differed from open to closed conformation, probably due to the difference between the folds of the oxyanion hole from open to closed state. Similarly, we note that the open form has a slightly less compact catalytic site than that of the closed form at 310 K (Fig. 6a), probably due to enhanced flexibility of the active site in the open conformation. Notwithstanding such natural discrepancies between closed and open state measurements, this analysis led us to reach similar conclusions with RMSD, RMSF, salt bridge, and structural visualizations. As such, the triad distances and radius of gyration of the catalytic machinery were found to be unchanged in the presence of non-polar organic solvent at 450 K while they became largely separated in the presence polar organic solvents (Fig. 6b and Fig. S7), suggesting an unfolding in the presence of polar solvent. Given these comprehensive theoretical analyses, one would expect an improved structural stability of lipase in non-polar organic solvents than in water, particularly at elevated temperatures.
Experiments corroborate with the simulations: lipase thermostability is enhanced in non-polar solvents
To validate the insights gained from MD simulations, we experimentally studied the effect of different organic solvents on the lipase activity at varying concentrations and temperatures. For this purpose, BTL2 gene harboring plasmid was expressed in E. coli and purified (Fig. S8). Recombinant BTL2 exhibited its maximum activity in 2-propanol and ethanol. As such, the activity increased by 4.75 and 3.46-fold in 80% of 2-propanol and 70% of ethanol, respectively (Fig. 8a). These improvements were observed to be much less for acetone and methanol when compared with the rest of the polar solvents. BTL2 activity was increased by 1.66 and 1.72 folds in 40% methanol and 60% acetone, respectively. On the other hand, BTL2 activity was slightly decreased at high concentrations of ethanol and 2-propanol while high concentrations of acetone and methanol completely inactivated the enzyme. Such inhibitory effects of methanol and acetone, particularly at high concentration, are likely to occur due to decreased solubility of the BTL2 since these two solvents lower protein solubility and often used for protein precipitation [41]. Furthermore, similar inhibitory effects of acetone on lipases were previously reported that it replenished the activity of a thermostable lipase from isolated from Bacillus sp. at concentrations higher than (60% v/v) [42]. Notably, we observed that the Phe17 which is the sole residue that changes its conformation from open-to-close state by adapting an alternate side chain conformation (Δχ = 110°) [19] started to attain the closed conformation in the presence of methanol. On the other hand, in water and ethanol, the side chain of Phe17 maintained its open conformer (Fig. 7). Hence, such inhibitory effect of methanol can be resulted from the altered Phe17 conformation which blocks the entrance to the active site.
Generally, all the organic solvents were found to improve lipase activity until a specific concentration after which the activity either diminished or completely lost (Fig. 8a). Ethanol and 2-proponal have folded the lipase activity up to 5 and 3 folds respectively. On the other hand, non-polar solvents did not make such a significant increase in the activity. Enzymes conserve some level of flexibility particularly at their active site for substrate binding and catalysis [43, 44]. Thus, it should be noted that polar solvents might be more advantageous than non-polar solvents to maintain a level of flexibility at low temperatures where atomic fluctuations are naturally low. Parallel with this assertion, we observed that the polar solvents remarkably enhanced the lipase activity at ambient temperatures (Fig. 8a). Furthermore, lipases are unique enzymes that depart from conventional Michaelis-Menten kinetics owing to the fact that they are solved at the same phase with their substrates [45]. Hence, enhanced activity in organic solvents regardless of the polarity might be resulted from increased solubility of the substrates in organic solvents than in water. Toshi-Asakura et al. observed that organic solvents were found to stabilize hemoglobin at low concentrations, while the protein becomes denatured at high concentrations. Similar effects of organic solvents were reported also on sickle cell hemoglobin [46].
The behavior of lipase in non-polar solvents was also analyzed (Fig. 8a). Lipase activity increased by 2.78 and 1.73 folds in 70% cyclohexane and 50% toluene, respectively. However, a severe drop was noted at the high concentrations of non-polar solvents. These lowered activities could be due to the high stiffness of the BTL2 in these solvent which may hamper the enzymatic activity [47]. As observed for the 310 K simulations, the lipase structure regardless of the lid conformation maintained a rigid conformation in the non-polar organic solvents (Fig. 2a), (Fig. S2A), and (Fig. 3a). High stiffness in the lipase structure along with reduced solubility of the lipase in non-polar solvents might account for the diminished activity (Fig. 8a).
Aiming to understand the link between the organic solvents and thermostability, the residual activity was monitored upon incubation of the lipase in the presence of organic solvents at elevated temperature. It was noted that lipase restored higher activity when incubated in the presence of ethanol and cyclohexane than in water (Fig. 8b). Ethanol increased the lipase activity by 4.89 folds at 25%, but no activity was observed at higher concentrations (50% and 70%) (Fig. 8B). Such high stability in ethanol was anticipated by the 450 K simulations which showed that the catalytic machinery of lipase was maintained in the presence of ethanol (Fig. 6 and Fig. S6). One the other hand, heating up lipase at high concentrations of polar solvents such as ethanol and water led to over flexibility and thus may impart in inducing protein unfolding. Such high flexibility was clearly seen in the RMSF profiles of the backbone atoms in the polar solvents at high temperatures (Fig. 2b, Fig. S2A, and Fig. 4). In addition to the over flexibility of lipase in polar solvents, an increased diffusion of the solvent molecules into the protein hydrophobic core may contribute to the protein unfolding by disturbing its hydrophobic core.
Cyclohexane on the other side improved lipase activity by 2.5 and 2.8 folds at 50% and 70% (v/v), respectively, and no denaturation or in activation was observed even at such high concentrations. These observations are consistent with our high temperature simulation results. Lipase regardless of its conformation was found to be very stable over all the simulation period, and no unfolding was observed at such very elevated temperature (Fig. S1B). Enhanced thermal stability of proteins in non-polar solvents was reported to be due to the high rigidity of anhydrous solvents and their inert nature [48]. Such a very high rigidity was seen clearly in the RMSF and SASA, and Rg profiles of lipase in non-polar solvents at 450 K (Fig. 2b, Fig.3, Fig. S2B, and Fig.4a–b). Another reason for this high stability could be due to the burial of the peptide bonds and the other polar side chains inside the protein structure protecting the folded globular form from the hydrophobic environment [38]. Harpaz et al. analyzed the contribution of buried residues to the overall stability of 108 different proteins [49]. Their results showed that polar group burial contributes favorably to protein stability more than the burial of non-polar groups. This explains the high compactness of lipase catalytic machinery in non-polar solvents seen during our high temperature simulations.
Overall, our findings provide molecular insights that can be useful in the rational design of stable lipases and thus in the optimization of the conditions of lipases in industry. These findings can also be adapted to other lipases especially in the lipase family I.5 and different enzyme groups. In conclusion, this study is an effort to explain a unifying perspective to lipase stability at extreme conditions with a guidance for protein engineering studies.
Conclusions
The results obtained by this study shed light on the effect solvent polarity on the stability and the activity of thermoalkalophilic lipases. The study also reflects the utility of computational methods in understanding protein stability and activity at extreme conditions at the molecular level. Altogether, our findings can help in tailoring the solvent environments for lipase reactions to improve their implementations in industry. Future studies will be centered on understanding the stability mechanism of lipases in the non-conventional deep eutectic solvents (DES). Despite the growing number of studies utilizing DES in lipase reactions, the mechanism of lipase stability in this solvent is still elusive at the molecular level.
Data availability
All data will be provided in Excel files to anyone requesting it.
References
Björkling F, Godtfredsen SE, Kirk O (1991) The future impact of industrial lipases. Trends Biotechnol. 9:360–363
Bornscheuer U, Reif OW, Lausch R, Freitag R, Scheper T, Kolisis FN et al (1994) Lipase of pseudomonas cepacia for biotechnological purposes: purification, crystallization and characterization. Biochim. Biophys. Acta 1201:55–60
Mukherjee KD (1990) Lipase-catalyzed reactions for modification of fats and other lipids. Biocatalysis. 3:277–293
Sharma R, Chisti Y, Banerjee UC (2001) Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 19:627–662
Borgström, B., Brockman. H.L. Lipases, Elsevier Science Ltd, 1984
Jaeger K-E, Eggert T (2002) Lipases for biotechnology. Curr. Opin. Biotechnol. 13:390–397
Bisen PS, Sanodiya BS, Thakur GS, Baghel RK, Prasad GB (2010) Biodiesel production with special emphasis on lipase-catalyzed transesterification. Biotechnol. Lett. 32:1019–1030
Reetz MT (2002) Lipases as practical biocatalysts. Curr. Opin. Chem. Biol. 6:145–150
Cox, M.M., Nelson, D.L. Lehninger Principles of Biochemistry, WH Freeman, 2008
Klibanov AM (1989) Enzymatic catalysis in anhydrous organic solvents. Trends Biochem. Sci. 14:141–144
Koskinen AM (1996) Klibanov. Springer, A.M. Enzymatic reactions in organic media
Zaks A, Klibanov AM (1984) Enzymatic catalysis in organic media at 100 degrees C. Science. 224:1249–1251
Durmaz E, Kuyucak S, Sezerman UO (2013) Modifying the catalytic preference of tributyrin in Bacillus thermocatenulatus lipase through in-silico modeling of enzyme-substrate complex. Protein Engineering, Design & Selection 26:325–333
Timucin E, Cousido-Siah A, Mitschler A, Podjarny A, Sezerman OU (2016) Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5. Proteins. 84:129–142
Timucin E, Sezerman OU (2013) The conserved lid tryptophan, W211, potentiates thermostability and thermoactivity in bacterial thermoalkalophilic lipases. PLoS One 8:e85186
Timucin E, Sezerman OU (2015) Zinc modulates self-assembly of Bacillus thermocatenulatus lipase. Biochemistry. 54:3901–3910
Yukselen O, Timucin E, Sezerman U (2016) Predicting the impact of mutations on the specific activity of Bacillus thermocatenulatus lipase using a combined approach of docking and molecular dynamics. Journal of Molecular Recognition 29:466–475
Palomo JM, Penas MM, Fernandez-Lorente G, Mateo C, Pisabarro AG, Fernandez-Lafuente R et al (2003) Solid-phase handling of hydrophobins: immobilized hydrophobins as a new tool to study lipases. Biomacromolecules. 4:204–210
Carrasco-Lopez C, Godoy C, de Las Rivas B, Fernandez-Lorente G, Palomo JM, Guisan JM et al (2009) Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 284:4365–4372
Jeong ST, Kim HK, Kim SJ, Chi SW, Pan JG, Oh TK et al (2002) Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J. Biol. Chem. 277:17041–17047
Martinez L, Andrade R, Birgin EG, Martinez JM (2009) PACKMOL: a package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 30:2157–2164
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E et al (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802
Huang J, MacKerell Jr AD (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34:2135–2145
Brooks BR, Brooks 3rd CL, Mackerell Jr AD, Nilsson L, Petrella RJ, Roux B et al (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30:1545–1614
MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586–3616
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935
Pastor RW, Brooks BR, Szabo A (1988) An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65:1409–1419
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N· log (N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38 27-8
Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD (2006) Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 22:2695–2696
Schmidt-Dannert C, Rua ML, Atomi H, Schmid RD (1996) Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochim. Biophys. Acta 1301:105–114
Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254
Derewenda ZS, Sharp AM (1993) News from the interface: the molecular structures of triacylglyceride lipases. Trends Biochem. Sci. 18:20–25
Huang X, Zhou HX (2006) Similarity and difference in the unfolding of thermophilic and mesophilic cold shock proteins studied by molecular dynamics simulations. Biophys. J. 91:2451–2463
Merkley ED, Parson WW, Daggett V (2010) Temperature dependence of the flexibility of thermophilic and mesophilic flavoenzymes of the nitroreductase fold. Protein Eng Des Sel. 23:327–336
Purmonen M, Valjakka J, Takkinen K, Laitinen T, Rouvinen J (2007) Molecular dynamics studies on the thermostability of family 11 xylanases. Protein Eng Des Sel. 20:551–559
Pace CN, Trevino S, Prabhakaran E, Scholtz JM (2004) Protein structure, stability and solubility in water and other solvents. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 359:1225–1234 discussion 34-5
Ahern TJ, Casal JI, Petsko GA, Klibanov AM (1987) Control of oligomeric enzyme thermostability by protein engineering. Proc. Natl. Acad. Sci. U. S. A. 84:675–679
Basu S, Biswas P (2018) Salt-bridge dynamics in intrinsically disordered proteins: a trade-off between electrostatic interactions and structural flexibility. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1866:624–641
Fic E, Kedracka-Krok S, Jankowska U, Pirog A, Dziedzicka-Wasylewska M (2010) Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 31:3573–3579
Sugihara A, Tani T, Tominaga Y (1991) Purification and characterization of a novel thermostable lipase from Bacillus sp. J. Biochem. 109:211–216
Kuntz Jr ID, Kauzmann W (1974) Hydration of proteins and polypeptides. Adv. Protein Chem. 28:239–345
Rupley JA, Careri G (1991) Protein hydration and function. Adv. Protein Chem. 41:37–172
Nini L, Sarda L, Comeau LC, Boitard E, Dubes JP, Chahinian H (2001) Lipase-catalysed hydrolysis of short-chain substrates in solution and in emulsion: a kinetic study. Biochim. Biophys. Acta 1534:34–44
Asakura T, Adachi K, Schwartz E (1978) Stabilizing effect of various organic solvents on protein. J. Biol. Chem. 253:6423–6425
Schmitke JL, Wescott CR, Klibanov AM (1996) The mechanistic dissection of the plunge in enzymatic activity upon transition from water to anhydrous solvents. J. Am. Chem. Soc. 118:3360–3365
Klibanov AM (2001) Improving enzymes by using them in organic solvents. Nature. 409:241–246
Harpaz Y, Gerstein M, Chothia C (1994) Volume changes on protein folding. Structure. 2:641–649
Funding
This work supported by The Scientific and Technological Research Council of Turkey and The National Research Council of Italy (TUBITAK grant no. 215Z712). The numerical calculations reported in this paper were partially performed at TUBITAK ULAKBIM, High Performance and Grid Computing Center (TRUBA resources).
Author information
Authors and Affiliations
Contributions
M.S. performed the experimental and in silico studies and wrote the manuscript. E.T. supervised both of the experimental and in silico work and wrote the manuscript. A.V. co-supervised in silico studies and reviewed the manuscript. O.U.S. reviewed the manuscript and offered advice in silico work.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1830 kb)
Rights and permissions
About this article
Cite this article
Shehata, M., Timucin, E., Venturini, A. et al. Understanding thermal and organic solvent stability of thermoalkalophilic lipases: insights from computational predictions and experiments. J Mol Model 26, 122 (2020). https://doi.org/10.1007/s00894-020-04396-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-020-04396-3