Abstract
Mutations in Cu/Zn superoxide dismutase 1 (SOD1) protein are a major cause of the devastating neurodegenerative disorder Amyotrophic lateral sclerosis. Evidence suggests that SOD1 functions as a free radical scavenger in humans. However, neither the mechanism nor a cure for this neurodegenerative disease are yet known. In the present study, we explored the effect of mutations on the mechanistic action on the Zn binding loop of SOD1 through discrete molecular dynamics. The results were analyzed in detail using statistical potential (BACH) to find the mutant structures having the least potential energy. Subsequently, we studied the impact of those mutations on metal ions bound in SOD1 using the program Check My Metal. Remarkably, our results recognized certain mutants, viz. His80Arg and Asp83Gly, that were more damaging to the Zn binding loop than all other mutants, leading to a loss of Zn binding with altered coordination of the Zn ion. Furthermore, the conformational stability, compactness, and secondary structural alteration of the His80Arg and Asp83Gly mutants were monitored using distinct parameters. Hence, at low computational expense, our study provides helpful insight into this emergent neurodegenerative disorder affecting mankind.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metal ions in proteins are essential, and have profound effects on protein stability and biological activity. Certain metals, such as sodium, magnesium, calcium and potassium, exist in the human body in significant amounts, while the other transition metals (Cu, Zn, Fe, Co, Mn and Ni) are present at low concentration [1, 2]. The superoxide dismutase (SOD1) enzyme belongs to one such metalloproteinase family present in the cytosol, mitochondria and nuclear membrane that functions by scavenging the free superoxide radicals generated during normal biochemical metabolism [3]. Monomeric SOD1 comprises 153 polypeptides bound with one Cu and Zn ion. The structure of SOD1 is organized as eight beta-barrel strands and two functionally important loops, i.e., a Zn loop (49–82) and an electrostatic loop (121–142). The metal ions (Cu and Zn) present in SOD1 are highly coordinated with their binding residues. The Cu ion is bound by a group of His residues (46, 48, 63 and 120), while the Zn ion is bound to His (63, 71, 80) and Asp 83 [4–6]. Thus, the Cu and Zn ions are essential for dismutase activity and the structural integrity of SOD1, respectively, i.e., the coordination of metal ions (Cu and Zn) in SOD1 supports both biological functioning and protein folding [7–10]. Mutation of SOD1 can give rise to the fatal neurodegenerative disorder, familial amyotrophic lateral sclerosis (FALS) [11, 12]. Mutations in SOD1 have been found over the entire length of protein, leading to more than 160 prominent mutations that could instigate FALS in humans [13, 14].
In the present study, we focused on the prominent missense mutations in the Zn binding loop residues (49–82) of SOD1 reported in the ALS database [14], since loss of the Zn ion in SOD1 could lead to ALS, by altering the protein-folding pattern and affecting catalytic activity. Further, the disparity in Zn binding affects the stability of SOD1, as it is crucial for retaining the structure of SOD1. Moreover, SOD1 with a Cu ion becomes catalytically inactive without the presence of a Zn ion. In addition, experimental studies have indicated that the SOD1 protein without the Zn ion has a distorted Zn and electrostatic loop, while loss of the Cu ion retains stability [15, 16]. Thus, we addressed this aspect using computational techniques to find the most lethal mutant that could lead to loss of Zn binding and, therefore, SOD1 stability, through various parameters. Hence, our report contributes to the search for mutational effects on SOD1 protein via discrete molecular dynamics
Materials and methods
Structure retrieval and geometry optimization
The X-ray crystal structure of native SOD1 was obtained from PDB (2V0A [A]) at a resolution of 1.15 Å [17]. The Zn binding loop mutants (Table 1) reported in the ALS database were retrieved for our study and modeled using Swiss PDB viewer with the native structure as a template. The native and mutant structures were energy minimized, using GROMACS v5.0.4 [18].
Effect of mutation on protein stability
The stability of Zn loop mutants was predicted via the program iStable, which utilizes the support vector machine (SVM) for stability predictions upon a single point mutation in protein sequence [19]. The result obtained from iStable integrates the prediction from various other programs such as CUPSAT, MUpro, PoPMuSiC, i-Mutant 2.0 and AUTO-MUTE. Further, we incorporated the PREDICT-SNP program [20], which is a consensus classifier for predicting mutant effects on SOD1 function. The program combines the results from different tools (SIFT, SNAP, PhD-SNP, PolyPhen-1, PolyPhen-2 and MAPP), and predicts the consolidated results along with a confidence score.
Discrete molecular dynamics
We performed dynamic analyses for native and mutant SOD1 using discrete molecular dynamics (DMD) simulation, a distinct molecular dynamics that uses a discrete energetic potential for pairwise interaction modeled with discontinuous functions [21–23]. An atomistic DMD force field was used in this study. The united atom model was used to represent a protein model in which polar hydrogen atoms and heavy atoms were modeled. Bonded interactions comprise covalent bonds, bond angles and dihedrals. The non-bonded interactions include van der Waals, solvation, and environment-dependent hydrogen bond interactions. The Lazaridis-Karplus implicit solvation model was used to model the solvated energy with fully solvated conformations as the reference state. Hydrogen bond interactions were modeled using reaction-like algorithms. Screened charge–charge interactions were modeled, using Debye-Hückel approximation, by setting Debye length to ∼10 Å. DMD simulation was performed at constant volume and periodic boundary conditions. An Anderson thermostat was used to maintain a constant temperature of 300 K throughout the DMD simulation for native and mutant SOD1. The Zn and Cu ions were added independently before performing simulation for native and mutant SOD1. The dynamic simulation was performed at timeunits (tu) of 1 × 105 for native and mutant SOD1, respectively. In this study, we performed DMD simulation for native and mutant SOD1 independently. The obtained trajectories were analyzed using the Bayesian Analysis Conformation Hunt (BACH) program [24], which reveals the potential score of native and mutant SOD1 in locating the structures with the lowest potential score in terms of kilocalories per mole.
Zn coordination and ensemble analysis
The mutant structures obtained from the BACH program were subjected to Zn binding analysis, using the program Check My Metal (CMM). The CMM program uses the NEIGHBORHOOD algorithm, which recognizes and examines the coordinated geometry of metal ions present in SOD1 [25]. The coordination geometry of metal ions was assessed using different parameters [Occupancy, B-factor, Ligands, Valence, nVECSUM, Geometry, gRMSD(°) and Vacancy] with the threshold values obtained from datasets. Thus, the Occupancy and Ligands parameter in CMM program designates the metal occupancy and ligand composition for the first coordination sphere of the metals, respectively. Further, the Valence parameter specifies the individual bond valance present in the metal. The nVECSUM parameter postulates the sum vector of the valance vector of the metal. Finally, the Geometry, gRMSD(°) and the Vacancy parameters determines the three dimensional coordination of the ligands, geometrical deviations and the vacancy in the coordination space around the metals, individually. Furthermore, the results from the program were displayed in green, yellow and red colors denoting acceptable, borderline, and outlier values of metal coordination, respectively. In addition, the program also provided the distance distribution of the metal complex in comparison with the Cambridge structural database [26]. Therefore, an outcome from the CMM program with a red color indication in more than two parameters suggests an error in metal binding. Thereby, we detected the most lethal mutant, which could lead to loss of Zn binding. Further, the trajectory of the most lethal mutant structures was analyzed to determine the conformational stability, compactness and the secondary structural alterations in SOD1. Thus, the geometrical tools g_rms, g_gyrate, do_dssp present in the GROMACS [18] program were utilized in our study, respectively.
Results and discussion
Earlier studies on SOD1 indicated that the loss of Zn ion could lead to ALS [16]. In this study, using various computational tools, we analyzed the effect of mutation on the Zn binding loop in order to narrow down the lethal mutant that could cause the disparity in Zn binding, and affect protein structural activity.
Mutant stability and function prediction
Earlier studies have reported that a single point mutation could lead to changes in protein stability [27–29]. Thus, we monitored the effect of sequence level changes on mutant SOD1 stability using the program iStable. The results predicted results by the program are presented in Table 1. Notably, increased stability was found in some mutants, viz., Cys57Arg, Ser59Ile, Gly61Arg, Phe64Leu, Asn65Ser, His71Tyr, Asp76Tyr, and Asp76Val, while other mutants exhibited a loss of SOD1 stability. Further, examination of the effect of mutants on SOD1 function using the program PREDICT SNP (Table 1) revealed that all these mutants showed a deleterious effect on SOD1 function, except Leu67Pro and Leu67Arg, which exhibited a neutral effect on SOD1. Therefore, our screen identified mutants (Glu49Lys, Thr54Arg, Pro66Ala, Pro66Ser, Pro66Arg, Gly72Ser, Gly72Cys His80Arg, Asp83Gly, Leu84Phe, Leu84Val) having both decreased and deleterious effects on SOD1 stability and corresponding function.
Structural dynamic study
The dynamic behavior of the native protein and screened mutants was analyzed using atomistic DMD simulation to study the effect of mutation on metal binding. The dynamics simulation was performed with a tu of 1 x 105 for all the native and mutant SOD1 proteins. The ensembles of native and mutant protein conformation were analyzed using the BACH program. The results specified the structure of native and mutant SOD1 having the lowest potential energy (in kcal mol−1). Those structures were recovered from the trajectory and further analyses were performed.
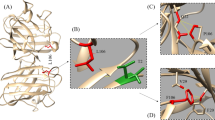
Coordination of metal ions
The CMM program was incorporated in our study to analyze the coordination of metal ions in native and mutant structures obtained from the BACH program. The results, showing acceptable, borderline and outlier values, are presented in Table 2. Initially, metal ion occupancy was determined in native and mutant SOD1. Subsequently, we determined ligand parameters designating similar and acceptable ligands (N4) for the Cu ion in the entire native and mutant structures. Notably, the ligands (O1N2) for Zn ion were found to be acceptable in all the mutants tested except His80Arg (O1N1) and Asp83Gly (N2). Further, the valence parameter endured acceptable, borderline outlier values for the native and mutant structures. In particular, the CMM indicated error in the valance value of the Zn ion for His80Arg and Asp83Gly, with values of 0.9 and 1, respectively. Progressively, the nVECSUM values were acceptable for Cu ion, though for Zn ion, which was found to be borderline and outlier. Consequently, the Geometry parameter specified square planar and tetrahedral geometry for the Cu ion, while distinct geometries were found for the Zn ion coordination. Furthermore, the gRMSD parameter indicated borderline and outlier values for the Cu and Zn ion, respectively. Moreover, the vacancy parameter clearly portrayed that the mutation had created a vacancy in Zn ion coordination in all the mutants tested. Specifically, Pro66Ala, His80Arg and Asp83Gly exhibited a greater vacancy with higher percentile. Likewise, CMM reports signified that the nVECSUM, Geometry and Vacancy parameters were perceived to decide potential error in metal binding, since these parameters stipulate the completeness of metal ion binding in SOD1 [25]. Hence, the above investigations inferred that His80Arg and Asp83Gly were the most destructive Zn binding mutants, exhibiting outlier values in the valance, nVECSUM, Geometry and Vacancy parameters as compared to other mutants.
Ensemble analysis
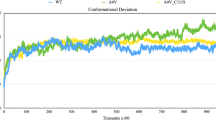
From the aforementioned report, the ensembles of native and mutant (His80Arg and Asp83Gly) were examined via distinct parameters in order to decipher the impact of the mutation on the structure of SOD1. Firstly, the root mean square deviation (RMSD) was processed for the c-alpha carbon atom of the native and mutant structures (Fig. 1a). From the overall examination, His80Arg and Asp83Gly achieved higher RMSD values (0.48 nm and 0.47 nm) than that of native SOD1 (0.46 nm). In this way, we also monitored the movement of the Zn binding loop in SOD1 via computing the c-alpha RMSD for the residues (49–82) within the native and mutant SOD1. Surprisingly, an increased RMSD was found in the mutant compared to the native protein (Fig. 1b). At first, Asp83Gly showed a higher RMSD with 0.5 nm, while identical deviation (0.4 nm) was found in the native protein and H80R, until 0.1x105 tu. From that point onwards, the motion of the Zn binding loop in the native differed between 0.4 nm and 0.5 nm, and focused on an RMSD of 0.43 nm while the deviation in His80Arg and Asp83Gly demonstrated an expanded RMSD of 0.6 nm and 0.55 nm, respectively, at the end of the simulation. Therefore, it was confirmed that His80Arg and Asp83Gly had radically changed the movement of the Zn loop, explaining the increased RMSD in mutant SOD1, when contrasted with that of native SOD1.
The conformational stability of SOD1 plotted using the Grace program obtained from discrete molecular dynamics (DMD). a Root mean square deviation (RMSD) of native (NT) and mutant (His80Arg and Asp83gly) over the entire trajectory. b Specific RMSD for the Zn binding loop residues of native (NT) and mutant (His80Arg and Asp83gly) SOD1
The compactness of SOD1 was also analyzed, utilizing the g_gyrate tool. The outcomes acquired were plotted employing the Xmgrace tool as delineated in Fig. 2. The graph revealed that His80Arg and Asp83Gly have a similar compactness (1.52 nm) from 0.2–0.5 × 105 tu, while the compactness of the native protein remained < 1.5 nm. Indeed, the compactness of native, His80Arg and Asp83Gly converged at 1.47 nm, 1.55 nm and 1.52 nm, respectively, at the end of the simulation. Additionally, the average compactness was computed, revealing that the value for the native protein (1.47 nm) had better compactness than mutants His80Arg and Asp83Gly, with 1.53 nm and 1.52 nm, respectively. Overall, it was found that mutation at residues 80 and 83 coincided with altered conformational motion and loss in compactness of mutant SOD1, compared to native SOD1. In addition, we also performed a simulation with the mutant Cys57Arg, which was found to have no significant impact on SOD1 structure. The outcomes were found to be roughly similar to those of the native (Table S1), thereby confirming that the simulation method chosen does not introduce statistical noise.
Secondary structure composition
Further study was carried out on the ensemble of native and mutant SOD1 to ascertain the percentage of secondary structure composition using the DSSP program. The outcomes (seeTable 3) uncovered the content of α-helix, β-sheets, others (turn, bend, β-bridge and coil) in the native and the mutant. Analysis of the outcomes revealed that the amount of α-helix was higher (6%) in native SOD1 than in the mutants (His80Arg and Asp83Gly with 5% and 1%, respectively). In contrast, the propensity to form β-sheets was found to be higher in mutant (41%) than native SOD1 (40%). Further, the content of other structures in the mutants (54% and 58%) was more prominent than in the native protein (54%). In this manner, the outcomes indicated that the change in α-helix content had prompted the expansion in the structure of β-sheets and others in mutant SOD1. Studies have revealed that β-sheet is a key factor in protein folding [30]; therefore, the alteration in β-sheet content could bring about protein misfolding, eventually prompting diseased states [31, 32]. The transformation of helix into β-sheets is the major consideration in formation of the amyloid fibrils linked directly to the increase in aggregation propensity in SOD1 [33–35]. Moreover, the results obtained from our study correlate with earlier reports indicating the loss of stability and the metal binding in SOD1 mutants [36–38]. Taken together, investigation of the above parameters signified that these mutants could be devastating, not only in distorting Zn binding but also in instigating stability loss and aggregation in SOD1, which ultimately results in a neurodegenerative disorder [39, 40].
Conclusions
The computational work introduced in this study outlined the preliminary screening of deleterious and destabilizing mutants in the Zn binding loop of SOD1, using PREDICTSNP and iStable programs individually. Alongside, we studied the presence of metal binding in native and mutants with reduced potential retrieved from DMD. Thus, the obtained native and mutant structures were subjected to the CMM program to determine the coordination of metal ions in SOD1 using distinct parameters. The results revealed that the mutation on His80 and Asp83 led to a drastic loss in Zn binding compared to other mutants. Further, we explored the impact of those mutations on SOD1 structure through various geometrical parameters in order to determine the stability compactness and the composition of secondary structure. Thus, analyses of the above-mentioned parameters indicated that these mutation led to the destabilization and a loss in compactness, with increased beta-sheet propensity, as compared to native SOD1. Overall, the results revealed the cause of Zn binding loss, destabilization and aggregation to mutation in SOD1 that can finally lead to the fatal neurodegenerative disorder fALS in humans. This study provides a perspective on the most deleterious and destabilizing mutants in the Zn binding loop of SOD1 by using computational studies to enlighten their effect on metal ion coordination, stability, compactness and secondary structure propensity.
References
Djinovic-Carugo K, Carugo O (2015) Structural biology of the lanthanides—mining rare earths in the Protein Data Bank. J Inorg Biochem 143:69–76. doi:10.1016/j.jinorgbio.2014.12.005
He W, Liang Z, Teng M, Niu L (2015) mFASD: a structure-based algorithm for discriminating different types of metal-binding sites. Bioinformatics 31:1938–1944. doi:10.1093/bioinformatics/btv044
Osredkar J (2011) Copper and zinc, biological role and significance of copper/zinc imbalance. J Clin Toxicol S3:001. doi:10.4172/2161-0495.S3-001
Valentine JS, Doucette PA, Zittin Potter S (2005) Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem 74:563–593. doi:10.1146/annurev.biochem.72.121801.161647
Durer ZAO, Cohlberg JA, Dinh P et al (2009) Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One 4:e5004. doi:10.1371/journal.pone.0005004
Milardi D, Pappalardo M, Grasso DM, Rosa CL (2010) Unveiling the unfolding pathway of FALS associated G37R SOD1 mutant: a computational study. Mol Biosyst 6:1032–1039. doi:10.1039/B918662J
Furukawa Y, O’Halloran TV (2006) Posttranslational modifications in Cu, Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid Redox Signal 8:847–867. doi:10.1089/ars.2006.8.847
Galaleldeen A, Strange RW, Whitson LJ et al (2009) Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch Biochem Biophys 492:40–47. doi:10.1016/j.abb.2009.09.020
Perry JJP, Shin DS, Getzoff ED, Tainer JA (2010) The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta 1804:245–262. doi:10.1016/j.bbapap.2009.11.004
Leinartaitė L, Saraboji K, Nordlund A et al (2010) Folding catalysis by transient coordination of Zn2+ to the Cu ligands of the ALS-associated enzyme Cu/Zn superoxide dismutase 1. J Am Chem Soc 132:13495–13504. doi:10.1021/ja1057136
Rosen DR, Siddique T, Patterson D et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62. doi:10.1038/362059a0
Culotta VC, Yang M, O’Halloran TV (2006) Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta 1763:747–758. doi:10.1016/j.bbamcr.2006.05.003
Lill CM, Abel O, Bertram L, Al-Chalabi A (2011) Keeping up with genetic discoveries in amyotrophic lateral sclerosis: the ALSoD and ALSGene databases. Amyotroph Lateral Scler 12:238–249. doi:10.3109/17482968.2011.584629
Wroe R, Wai-Ling Butler A, Andersen PM et al (2008) ALSOD: the Amyotrophic Lateral Sclerosis Online Database. Amyotroph Lateral Scler 9:249–250. doi:10.1080/17482960802146106
Nedd S, Redler RL, Proctor EA et al (2014) Cu, Zn-superoxide dismutase without Zn is folded but catalytically inactive. J Mol Biol 426:4112–4124. doi:10.1016/j.jmb.2014.07.016
Hilton JB, White AR, Crouch PJ (2015) Metal-deficient SOD1 in amyotrophic lateral sclerosis. J Mol Med (Berl) 93:481–487. doi:10.1007/s00109-015-1273-3
Berman HM, Westbrook J, Feng Z et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. doi:10.1021/ct700301q
Chen C-W, Lin J, Chu Y-W (2013) iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinformatics 14:S5. doi:10.1186/1471-2105-14-S2-S5
Bendl J, Stourac J, Salanda O et al (2014) PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol 10:e1003440. doi:10.1371/journal.pcbi.1003440
Shirvanyants D, Ding F, Tsao D et al (2012) Discrete molecular dynamics: an efficient and versatile simulation method for fine protein characterization. J Phys Chem B 116:8375–8382. doi:10.1021/jp2114576
Ding F, Dokholyan NV (2006) Emergence of protein fold families through rational design. PLoS Comput Biol 2:e85. doi:10.1371/journal.pcbi.0020085
Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI (1998) Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des 3:577–587. doi:10.1016/S1359-0278(98)00072-8
Cossio P, Granata D, Laio A et al (2012) A simple and efficient statistical potential for scoring ensembles of protein structures. Sci Rep 2:351. doi:10.1038/srep00351
Zheng H, Chordia MD, Cooper DR et al (2014) Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc 9:156–170. doi:10.1038/nprot.2013.172
Groom CR, Bruno IJ, Lightfoot MP, Ward SC (2016) The Cambridge structural database. Acta Crystallogr Sect B: Struct Sci Cryst Eng Mater 72:171–179. doi:10.1107/S2052520616003954
Tokuriki N, Tawfik DS (2009) Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19:596–604. doi:10.1016/j.sbi.2009.08.003
Beckerman M (2015) Fundamentals of neurodegeneration and protein misfolding disorders. Springer, Basel
Ye L, Wu Z, Eleftheriou M, Zhou R (2007) Single-mutation-induced stability loss in protein lysozyme. Biochem Soc Trans 35:1551–1557. doi:10.1042/BST0351551
Khandogin J, Brooks CL (2007) Linking folding with aggregation in Alzheimer’s β-amyloid peptides. Proc Natl Acad Sci USA 104:16880–16885. doi:10.1073/pnas.0703832104
Gross M (2000) Proteins that convert from alpha helix to beta sheet: implications for folding and disease. Curr Protein Pept Sci 1:339–347
Soto C (2003) Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci 4:49–60. doi:10.1038/nrn1007
Cerdà-Costa N, Esteras-Chopo A, Avilés FX et al (2007) Early kinetics of amyloid fibril formation reveals conformational reorganisation of initial aggregates. J Mol Biol 366:1351–1363. doi:10.1016/j.jmb.2006.12.007
Dong M, Li H, Hu D et al (2016) Molecular Dynamics Study on the Inhibition Mechanisms of Drugs CQ1–3 for Alzheimer Amyloid-β40 Aggregation Induced by Cu2+. ACS Chem Neurosci. doi:10.1021/acschemneuro.5b00343
Stefani M (2004) Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta 1739:5–25. doi:10.1016/j.bbadis.2004.08.004
Krishnan U, Son M, Rajendran B, Elliott JL (2006) Novel mutations that enhance or repress the aggregation potential of SOD1. Mol Cell Biochem 287:201–211. doi:10.1007/s11010-005-9112-4
Khare SD, Dokholyan NV (2006) Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci USA 103:3147–3152. doi:10.1073/pnas.0511266103
Khare SD, Caplow M, Dokholyan NV (2004) The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 101:15094–15099. doi:10.1073/pnas.0406650101
Meiering EM (2008) The threat of instability: neurodegeneration predicted by protein destabilization and aggregation propensity. PLoS Biol 6:e193. doi:10.1371/journal.pbio.0060193
Stevens JC, Chia R, Hendriks WT et al (2010) Modification of superoxide dismutase 1 (SOD1) properties by a GFP tag—implications for research into amyotrophic lateral sclerosis (ALS). PLoS One 5:e9541. doi:10.1371/journal.pone.0009541
Acknowledgment
The authors thank the management of Vellore Institute of Technology (VIT) University for providing the facilities and encouragement to carry out this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 28 kb)
Rights and permissions
About this article
Cite this article
Srinivasan, E., Sethumadhavan, R. & Rajasekaran, R. A theoretical study on Zn binding loop mutants instigating destabilization and metal binding loss in human SOD1 protein. J Mol Model 23, 103 (2017). https://doi.org/10.1007/s00894-017-3286-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3286-z