Abstract
The human epidermal growth factor receptor (HER) family plays a vital role in the development of resistance to treatments in estrogen receptor (ER)-positive breast cancer. This study investigated the correlation between protein and mRNA expressions of the HER family in ER-positive breast cancer. We dissected regions of invasive cancer from the frozen tissues of 34 patients with ER-positive breast cancer using laser-capture microdissection, followed by evaluation of the mRNA levels of the ER and HER family (EGFR, HER2, HER3, and HER4) using the quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay. In addition, we assessed the protein expressions of the ER and HER family using an immunohistochemical (IHC) assay. A significant correlation was observed between the ER protein and mRNA expressions. For HER2, HER3, and HER4, protein expressions significantly correlated with mRNA levels. We established significant correlations of the mRNA level between EGFR versus HER2, as well as EGFR versus HER3. Furthermore, a significant correlation of the mRNA level between HER2 and HER3 was illustrated. In conclusion, IHC evaluation may be reliable and representable for mRNA. Hence, this study established a marked correlation between the mRNA expressions of HER family members in patients with ER-positive breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Routine clinical practice has been using a combination of immunohistochemical (IHC) analyses, including estrogen receptor (ER), progesterone receptor, and human epidermal growth factor 2 (HER2) expression as well as the Ki67 labeling index, to select treatment of early-stage breast cancer [1]. In clinical practice, HER2 testing for protein expression and gene amplification is typically used to ascertain patients’ eligibility for HER2-targeted therapies using trastuzumab and contributes toward the improvement of the outcome of patients with breast cancer [2, 3]. HER2 is included in the human epidermal growth factor receptor (HER) family, which comprises four members, EGFR (HER1), HER2, HER3, and HER4. Recently, EGFR, HER3, and HER4 have garnered attention and have been proposed as targets of molecular-targeted therapy besides HER2 for breast cancer, such as lapatinib [4], neratinib [5], and pertuzumab [6].
ER-positive breast cancer is characterized by clinical and biological heterogeneity. Investigating tumor heterogeneity is a major challenge for precise biological findings and treatment selection for patients with ER-positive breast cancer [7, 8] Reportedly, ER is activated by signal cross talk between estrogen and growth factors, including the HER family, through transmembrane receptor phosphorylation [9, 10]. The cross talk between the ER and HER signaling pathways in ER-positive tumors is thought to be responsible for the lower response of these tumors to endocrine therapies and HER2-targeted therapies [9, 11]. However, in ER-positive breast cancer, the correlation for each member of the HER family remains unclear.
Hence, this study aimed to assess the protein and mRNA expressions of the HER family and compare these expressions for each member of the HER family in ER-positive breast cancer.
Patients and methods
Patient background and eligibility
In this study, we used frozen breast cancer samples obtained from 34 female patients with ER-positive invasive breast cancer diagnosed at the Saitama Cancer Center (Saitama, Japan) from 1998 to 2000. All patients underwent breast surgery without neoadjuvant treatment. Notably, all frozen samples were collected from surgical resection tissues.
This study was conducted according to the tenets of the Declaration of Helsinki, and the study protocol was approved by the Institutional Review Board of the Saitama Cancer Center (registration number: 379).
IHC evaluation of the ER and HER family
We examined ER and HER2 protein levels using the IHC assay. The sources of the primary antibodies were as follows: ER (1D5; DAKO, Denmark) and HER2 (HercepTest; DAKO, Denmark). We defined a positive or strong expression for ER according to the nuclear labeling index as expression levels ≥ 1%. In addition, the degree of staining for ER was assessed using the Allred score, in which the score was evaluated according to the proportion and intensity of positive cells [12]. Accordingly, HER2 testing using the IHC assay was assessed as follows: score 0, no staining or membrane staining in < 10% of tumor cells; 1+, faint perceptible membrane staining detected in ≥ 10% of tumor cells; 2+, weak to moderate complete membrane staining in ≥ 10% of tumor cells; and 3+, uniform, intense membrane staining in ≥ 10% of tumor cells [13]. In this study, we defined the sample with HER2 IHC ≥ 2 as HER2 positive.
We assessed EGFR, HER3, and HER4 protein levels using the IHC assay. For EGFR IHC, 4-µm paraffin sections were dewaxed, incubated with a proteolytic-induced epitope to retrieve the antigen for 7 min at room temperature (RT), and incubated with anti-human EGFR mouse monoclonal antibody (clone 2-18C9, DAKO) (×1) overnight at 4 °C. For HER3 IHC, dewaxed sections were incubated in high-pH antigen retrieval reagent for 40 min at 97 °C and incubated with anti-human HER3 mouse monoclonal antibody (clone DAK-H3-IC, DAKO) (×50) for 20 min at RT. For HER4 IHC, dewaxed sections were autoclaved in citrate buffer for 15 min and incubated with anti-human c-erb-4 rabbit polyclonal antibody (clone N/A, DAKO) (×50) for 60 min at RT. Subsequently, sections were incubated with horseradish peroxidase-conjugated polymer (Envision detection system, DAKO) and detected using DAB. All IHC procedures were performed by one technician under the supervision of one pathologist. In the IHC evaluation of the EGFR and HER3 expression, we evaluated the grades of membrane staining using the HER2 criteria. Samples with an IHC score ≥ 1 were assessed as positive. In the HER4 IHC assessment, we evaluated the cytoplasmic staining of cells and classified the samples stained ≥ 1% of cells as positive.
Laser-capture microdissection
We performed laser microdissection to precisely evaluate the mRNA expression in breast adenocarcinoma cells and the surrounding stroma. In addition, we examined the available histological sections using 8-μm sliced optimal cutting temperature compound-embedded frozen samples stained with cresyl violet to select the deepest invasive section from the tumor. Subsequently, cells were transferred from the slide onto the caps using PALM MicroBeam IV (Carl Zeiss Microscopy). Finally, the total microdissected area was adjusted approximately to 5 × 106 μm2 for each sample.
Quantitative real-time reverse transcription polymerase chain reaction assay
We extracted total RNA from the microdissected samples using the RNeasy Plus Micro Kit (Qiagen) according to the manufacturer’s instructions. The extracted RNA was converted to cDNA using a QuantiTect Reverse Transcription Kit (Qiagen). In addition, all transcripts were quantified by TaqMan polymerase chain reaction (PCR) using the Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technologies) and a Step One Real-Time PCR System (Applied Biosystems Inc.). Samples with a mean of the triplicate cycle threshold (CT) values for β-actin as the housekeeping gene ≤ 33 (t) were determined eligible for further investigation. Of note, the ΔCT value of the target gene (ΔCT = mean CT target gene − mean CT housekeeping gene) was defined as the relative mRNA expression level. Table 1 presents the sequences of gene-specific PCR primes and probes for ER, EGFR, HER2, HER3, HER4, and β-actin.
Statistical analysis
For ER, the comparisons between protein expression using the Allred score and mRNA expression using the ΔCT value were assessed using the Kruskal–Wallis test. For the HER family, the correlation of the protein expression using the IHC scoring and the mRNA expression using the ΔCT value was assessed using the Mann–Whitney U test. In addition, the correlation between each mRNA expression (ΔCT value) of the HER family was assessed using Spearman’s rank correlation test. The relationship between HER family expression and various clinicopathological factors was analyzed using the Chi-squared and Fisher’s exact tests. We defined P < 0.05 as statistically significant in this study. All statistical analyses were conducted using the GraphPad Prism 7.03 (GraphPad Software Inc.) and SPSS v24.0 (IBM Corp.) software.
Results
Patient and tumor characteristics
Table 2 summarizes the characteristics of patients and tumors in this study. The median age of the 34 patients was 51 years (age range 41–84 years). At the initial surgical point, 17 patients reported being in the postmenopausal stage. The pathological tumor sizes (pT) were as follows: pT1, 17 patients (50.0%); pT2, 7 patients (20.6%); pT3, 2 patients (5.9%); and pT4, 8 patients (23.5%). Furthermore, the pathological lymph node status (pN) was as follows: pN0, 15 patients (44.1%); pN1, 8 patients (23.5%); pN2, 5 patients (14.7%); and pN3, 6 patients (17.6%). Twenty patients (58.8%) had breast-conserving surgery, whereas 14 patients (41.2%) had mastectomy. In addition, 19 patients (55.9%) underwent axillary lymph node dissection.
Distribution of patients stratified according to the HER status
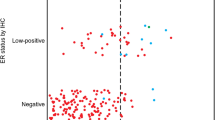
The distribution of patients stratified according to the IHC expression of EGFR was as follows: IHC 0, 33 patients (97.1%); and IHC 1+, 1 patient (2.9%). The distribution of patients stratified according to the HER2 IHC expression was as follows: IHC 0, 21 patients (61.8%); IHC 1+, 8 patients (23.5%); IHC 2+, 2 patients (5.9%); and IHC 3+, 3 patients (8.8%). In addition, the distribution of patients stratified according to the HER3 IHC expression was as follows: IHC 0, 27 patients (79.4%); IHC 1+, 3 patients (8.8%); and IHC 2+, 4 patients (11.8%). Furthermore, the distribution of patients stratified according to the HER4 IHC expression was as follows: IHC negative, 17 patients (50.0%); and IHC positive, 17 patients (50.0%). In this study, IHC-positive samples were only 1 tumor (2.9%) for EGFR, 5 tumors (14.7%) for HER2, 7 tumors (20.6%) for HER3, and 17 tumors (50.0%) for HER4 (Table 3). Figure 1 shows IHC-positive cases for each HER family member. The median relative expression of mRNA encoding using ΔCT values was as follows: ER, − 1.08 (2.86 to − 3.17); EGFR, 6.17 (9.73 to 2.18); HER2, − 0.70 (4.15 to − 3.61); HER3, 0.43 (3.42 to − 1.60); and HER4, 2.26 (6.28 to − 1.11; Table 3). The comparison between the HER family expressions and clinicopathological factors is shown in Table 4.
Concordance between the mRNA and protein expressions for the ER and HER family
The number of samples available for analysis was 34 (100%) for ER, 25 (73.5%) for EGFR, 33 (97.1%) for HER2, 34 (100%) for HER3, and 33 (97.1%) for HER4. We observed a significant correlation between the ER protein expression and the ER mRNA status (P = 0.031). For HER2, HER3, and HER4, protein expressions significantly correlated with mRNA levels (HER2: P = 0.009; HER3: P = 0.035; HER4: P = 0.011). For EGFR, we did not perform a statistical analysis because only 1 patient had EGFR IHC-positive tumor (Table 3).
Correlation among mRNA expressions of the HER family
For the HER3 mRNA expression, we observed a significant correlation with the ER mRNA expression (r = 0.35; P = 0.040), but not with other members (Fig. 2). In addition, we determined a positive correlation between the EGFR and HER2 mRNA expression levels (r = 0.60; P = 0.002), as well as between the EGFR and HER3 mRNA values (r = 0.42; P = 0.039). Furthermore, we observed a significant relationship between the HER2 and HER3 mRNA expression levels (r = 0.37; P = 0.036), respectively. Figure 3 illustrates scatter plots of the mRNA expression values of different HER family members.
Discussion
Approximately, 70% of breast cancer cases were ER positive [14]. ER controls the expression of various genes and proteins through genomic and non-genomic pathways. In the non-genomic pathway of ER, growth factors on the cellular membrane (including the HER family) primarily mediate cancer cell proliferation [9, 13, 15]. This pathway may be related to the development of resistance to treatments and outcome in patients with ER-positive breast cancer [9]. A recent clinical trial indicated that neratinib—a small-molecule tyrosine kinase inhibitor of HER1, HER2, and HER4—exhibited good efficacy in patients with hormone receptor-positive and HER2-positive breast cancer [5]. In addition, Pawlowski et al. reported an inverse correlation between the EGFR/HER2 mRNA expressions and ER mRNA expression. Conversely, the HER3 mRNA expression positively correlated with the ER mRNA expression in all subtypes of breast cancer [15]. Our present study revealed a marked correlation between the ER and HER3 mRNA expressions in ER-positive breast cancer. Although the kinase domain of HER3 lacks the catalytic function, HER3 is transphosphorylated and acts as a potent signaling platform through the heterodimer with other HER family members [16, 17]. The current study suggested a positive correlation between the HER3 and EGFR/HER2 mRNA expressions. Reportedly, the upregulation of the non-genomic pathway through HER3 with EGFR and HER2 may play a vital role in the regulation of cell growth, differentiation, and other factors involved in tumor formation in ER-positive breast cancer [18]. Nevertheless, further research involving large breast cancer cohorts is essential to elucidate the correlation between the HER family-related pathway and resistance to endocrine therapy, as well as disease progression in patients with ER-positive breast cancer.
Tumor heterogeneity is considered to be correlated with the response to molecular-targeted treatments including endocrine therapy and trastuzumab in breast cancer. In addition, it may be associated with poor prognosis in patients with breast cancer [19, 20]. Notably, refining and deciphering the molecular mechanisms which control tumor heterogeneity in breast cancer are imperative to attain a personalized approach to the management of patients with breast cancer. It is speculated that the isolation of mRNA from cancer cells using laser-capture microdissection may enhance the sensitivity and accuracy of assessment for tumor heterogeneity using the quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay approach. Hofmann et al. reported that qRT-PCR is entirely consistent with IHC using laser-capture microdissection for HER2 evaluation [21]. In this study, IHC assessment was shown to be reliable and representable for the mRNA in the evaluation of the ER and HER family expression. Furthermore, this study suggested that the combination of qRT-PCR and laser-capture microdissection may represent a valid alternative method for assessing the expression of the ER and HER family. Nevertheless, further studies are warranted to investigate opportunities for the development of a quantitative evaluation approach for ER-positive breast cancer with tumor heterogeneity.
Recent efforts have been focused on RNA-targeting molecular drugs such as small interfering RNAs (siRNA), microRNAs, and single-stranded antisense oligonucleotides (AONs) [22,23,24]. Numerous clinical trials of RNA-targeting molecular drugs have been conducted in various types of cancer [22]. However, in breast cancers, almost all clinical trials failed to show improved clinical outcomes and further development is difficult at the present time [22]. Previous in vitro and in vivo studies reported that a novel siRNA agent silenced EGFR and HER2 transcription, and inhibited breast cancer cell growth and invasiveness [25, 26]. Further biological and translational studies investigating the ability of new RNA-targeting molecular agents to down regulate the mRNA expression of the HER family are warranted.
In conclusion, the expression grades of protein in paraffin-embedded specimens correlated well with mRNA expressions of HER2, HER3, and HER4 obtained from frozen tissues of patients with ER-positive breast cancer. For EGFR, the IHC-positive rate was very low without correlation between the expressions of protein and mRNA. Hence, this study established a marked correlation among the mRNA expressions of each HER family member in patients with ER-positive breast cancer.
References
Curigliano G, Burstein HJ, Winer PE, Gnant M, Dubsky P, Loibl S, Colleoni M, Regan MM, Piccart-Gebhart M, Senn HJ, Thürlimann B, St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2017, André F, Baselga J, Bergh J, Bonnefoi H, Brucker YS, Cardoso F, Carey L, Ciruelos E, Cuzick J, Denkert C, Di Leo A, Ejlertsen B, Francis P, Galimberti V, Garber J, Gulluoglu B, Goodwin P, Harbeck N, Hayes DF, Huang CS, Huober J, Hussein K, Jassem J, Jiang Z, Karlsson P, Morrow M, Orecchia R, Osborne KC, Pagani O, Partridge AH, Pritchard K, Ro J, Rutgers EJT, Sedlmayer F, Semiglazov V, Shao Z, Smith I, Toi M, Tutt A, Viale G, Watanabe T, Whelan TJ, Xu B (2017) De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol 28:1700–1712
Seidman AD, Fornier MN, Esteva FJ, Tan L, Kaptain S, Bach A, Panageas KS, Arroyo C, Valero V, Currie V (2001) Weekly trastuzumab and paclitaxel therapy for metastatic breast cancer with analysis of efficacy by HER2 immunophenotype and gene amplification. J Clin Oncol 19:2587–2595
Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, Goldhirsch A, Untch M, Mariani G, Baselga J (2007) 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet 369:29–36
Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, Skarlos D, Campone M, Davidson N, Berger M, Oliva C, Rubin SD, Stein S, Cameron D (2006) Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med 355:2733–2743
Martin M, Holmes FA, Ejlertsen B, Delaloge S, Moy B, Iwata H, von Minckwitz G, Chia SKL, Mansi J, Barrios CH, Gnant M, Tomašević Z, Denduluri N, Šeparović R, Gokmen E, Bashford A, Ruiz Borrego M, Kim SB, Jakobsen EH, Ciceniene A, Inoue K, Overkamp F, Heijns JB, Armstrong AC, Link JS, Joy AA, Bryce R, Wong A, Moran S, Yao B, Xu F, Auerbach A, Buyse M, Chan A, ExteNET Study Group (2017) Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 18:1688–1700
Baselga J, Cortés J, Kim SB, Im SA, Hegg R, Im YH, Roman L, Pedrini JL, Pienkowski T, Knott A, Clark E, Benyunes MC, Ross G, Swain SM, CLEOPATRA Study Group (2012) Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 366:109–119
Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, Tsui DW, Liu B, Dawson SJ, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah S, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Børresen-Dale AL, Earl HM, Pharoah PD, Ross MT, Aparicio S, Caldas C (2016) The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 7:11479
Haricharan S, Bainbridge MN, Scheet P, Brown PH (2014) Somatic mutation load of estrogen receptor-positive breast tumors predicts overall survival: an analysis of genome sequence data. Breast Cancer Res Treat 146:211–220
Hayashi S, Yamaguchi Y (2008) Estrogen signaling pathway and hormonal therapy. Breast Cancer 15:256–261
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J (2017) Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet 62:15–24
Kurozumi S, Inoue K, Takei H, Matsumoto H, Kurosumi M, Horiguchi J, Takeyoshi I, Oyama T (2015) ER, PgR, Ki67, p27(Kip1), and histological grade as predictors of pathological complete response in patients with HER2-positive breast cancer receiving neoadjuvant chemotherapy using taxanes followed by fluorouracil, epirubicin, and cyclophosphamide concomitant with trastuzumab. BMC Cancer 15:622
Allred DC, Harvey JM, Berardo M, Clark GM (1998) Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 11:155–168
Kurozumi S, Matsumoto H, Hayashi Y, Tozuka K, Inoue K, Horiguchi J, Takeyoshi I, Oyama T, Kurosumi M (2017) Power of PgR expression as a prognostic factor for ER-positive/HER2-negative breast cancer patients at intermediate risk classified by the Ki67 labeling index. BMC Cancer 17:354
Johnston SR, Dowsett M (2003) Aromatase inhibitors for breast cancer: lessons from the laboratory. Nat Rev Cancer 3:821–831
Pawlowski V, Révillion F, Hebbar M, Hornez L, Peyrat JP (2000) Prognostic value of the type I growth factor receptors in a large series of human primary breast cancers quantified with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res 6:4217–4225
Citri A, Skaria KB, Yarden Y (2003) The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res 284:54–65
Park YH, Jung HA, Choi MK, Chang W, Choi YL, Do IG, Ahn JS, Im YH (2014) Role of HER3 expression and PTEN loss in patients with HER2-overexpressing metastatic breast cancer (MBC) who received taxane plus trastuzumab treatment. Br J Cancer 110:384–391
Koutras AK, Kalogeras KT, Dimopoulos MA, Wirtz RM, Dafni U, Briasoulis E, Pectasides D, Gogas H, Christodoulou C, Aravantinos G, Zografos G (2008) Evaluation of the prognostic and predictive value of HER family mRNA expression in high-risk early breast cancer: a Hellenic Cooperative Oncology Group (HeCOG) study. Br J Cancer 99:1775
Lindström LS, Yau C, Czene K, Thompson CK, Hoadley KA, Van’t Veer LJ, Balassanian R, Bishop JW, Carpenter PM, Chen YY, Datnow B, Hasteh F, Krings G, Lin F, Zhang Y, Nordenskjöld B, Stål O, Benz CC, Fornander T, Borowsky AD, Esserman LJ, STO Trialists Group (2018) Intratumor heterogeneity of the estrogen receptor and the long-term risk of fatal breast cancer. J Natl Cancer Inst 110:726–733
Kurozumi S, Padilla M, Kurosumi M, Matsumoto H, Inoue K, Horiguchi J, Takeyoshi I, Oyama T, Ranger-Moore J, Allred DC, Dennis E, Nitta H (2016) HER2 intratumoral heterogeneity analyses by concurrent HER2 gene and protein assessment for the prognosis of HER2 negative invasive breast cancer patients. Breast Cancer Res Treat 158:99–111
Hofmann E, Seeboeck R, Jacobi N, Obrist P, Huter S, Klein C, Oender K, Wiesner C, Hundsberger H, Eger A (2016) The combinatorial approach of laser-captured microdissection and reverse transcription quantitative polymerase chain reaction accurately determines HER2 status in breast cancer. Biomark Res 4:8
Barata P, Sood AK, Hong DS (2016) RNA-targeted therapeutics in cancer clinical trials: Current status and future directions. Cancer Treat Rev 50:35–47
Moreno PM, Pêgo AP (2014) Therapeutic antisense oligonucleotides against cancer: hurdling to the clinic. Front Chem 2:87
MacLeod AR, Crooke ST (2017) RNA therapeutics in oncology: advances, challenges, and future directions. J Clin Pharmacol 57:S43–S59
Gu S, Hu Z, Ngamcherdtrakul W, Castro DJ, Morry J, Reda MM, Gray JW, Yantasee W (2016) Therapeutic siRNA for drug-resistant HER2-positive breast cancer. Oncotarget 7:14727–14741
Tiash S, Kamaruzman NIB, Chowdhury EH (2017) Carbonate apatite nanoparticles carry siRNA(s) targeting growth factor receptor genes egfr1 and erbb2 to regress mouse breast tumor. Drug Deliv 24:1721–1730
Acknowledgements
We gratefully acknowledge the work of our research technicians (Akiyo Yamashita and Kumiko Sudo) at the Research Institute for Clinical Oncology and Department of Pathology in Saitama Cancer Center. We would like to thank Enago (http://www.enago.jp) for the English language review.
Funding
There is no funding to declare.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
This study was conducted according to the tenets of the Declaration of Helsinki and the study protocol was approved by the Institutional Review Board of Saitama Cancer Center (registration number: 379).
Rights and permissions
About this article
Cite this article
Kurozumi, S., Yamaguchi, Y., Matsumoto, H. et al. Comparing protein and mRNA expressions of the human epidermal growth factor receptor family in estrogen receptor-positive breast cancer. Med Mol Morphol 52, 90–98 (2019). https://doi.org/10.1007/s00795-018-0206-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00795-018-0206-y