Abstract
Larvae of the African midge Polypedilum vanderplanki show extreme desiccation tolerance, known as anhydrobiosis. Recently, the cultured cell line Pv11 was derived from this species; Pv11 cells can be preserved in the dry state for over 6 months and retain their proliferation potential. Here, we attempted to expand the use of Pv11 cells as a model to investigate the mechanisms underlying anhydrobiosis in P. vanderplanki. A newly developed vector comprising a constitutive promoter for the PvGapdh gene allowed the expression of exogenous proteins in Pv11 cells. Using this vector, a stable Pv11 cell line expressing green fluorescence protein (GFP) was established and retained desiccation tolerance. Gene silencing with GFP-specific siRNAs significantly suppressed GFP expression to approximately 7.5–34.6% of that in the non-siRNA-transfected GFP stable line. Establishment of these functional assays will enable Pv11 cells to be utilized as an effective tool to investigate the molecular mechanisms underlying anhydrobiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anhydrobiosis is a striking example of extreme desiccation tolerance. Anhydrobiotic organisms can tolerate almost complete dehydration, with their metabolic activity dropping to undetectable levels. If organisms enter anhydrobiosis, they are in a temporary state of apparent death, but they are not dead. In other words, they are still alive, despite a lack of internal water, but they show no visible signs of life. Once anhydrobiotic organisms are rehydrated, they resume active life and normal development. This phenomenon can be observed in many taxa, ranging from unicellular organisms to higher invertebrates and plants: examples are bacteria, yeast, fungal spores, protozoa, nematodes, bdelloid rotifers, tardigrades, and cysts of the brine shrimp (Watanabe 2006).
The sleeping chironomid Polypedilum vanderplanki, which inhabits semi-arid regions in Africa, is the largest animal anhydrobiote. During the process of anhydrobiosis induction by dehydration, the larvae accumulate trehalose (Watanabe et al. 2003) and late embryogenesis abundant (LEA) proteins (Kikawada et al. 2006; Cornette et al. 2010; Hatanaka et al. 2013), which are believed to play important roles in protecting biological components, such as proteins and cellular membranes from irreversible denaturation. Trehalose and LEA proteins are essential but insufficient for the successful induction of anhydrobiosis, as demonstrated in nematodes (Erkut et al. 2011) and P. vanderplanki (Sakurai et al. 2008; Shimizu et al. 2010). In fact, a mammalian cell line loaded with trehalose and expressing LEA proteins showed desiccation tolerance, but only to flash dehydration (Li et al. 2012). Therefore, other essential factors must be involved in the successful induction of anhydrobiosis.
Recently, the draft genome sequence of P. vanderplanki has been sequenced (Gusev et al. 2014). The genome and transcriptome project for P. vanderplanki revealed that some paralogous clustered genes encoding protective proteins or proteins involved in repair processes are likely to be involved in anhydrobiosis (Cornette et al. 2010; Gusev et al. 2014). However, this is just a prediction based on orthological similarities. Indeed, most of the predicted anhydrobiosis-related genes and the deduced proteins have not been characterized genetically or biochemically. Therefore, we need experimental evidence to confirm the contribution of these candidate genes to anhydrobiosis.
Pv11, a cultured cell line derived from the embryo of P. vanderplanki, displays extreme desiccation tolerance (Nakahara et al. 2010). Pv11 cells can be stored in a dry state at room temperature for up to 251 days, while retaining proliferation potential (Watanabe et al. 2016). Although genetic engineering applied to Pv11 cells should greatly improve our understanding of the molecular mechanisms underlying anhydrobiosis, efficient molecular engineering tools have not been developed for Pv11 cells. Indeed, in our experience, commercially available insect gene expression systems are not effective in Pv11 cells. Hence, we attempted to develop gene manipulation techniques to expand the usefulness of Pv11 cells as a tool for examining the mechanisms of anhydrobiosis in P. vanderplanki.
Materials and methods
Cell culture
Pv11 cells and their stable GFP-expressing derivatives (Pv11-KH cells) were cultured at 25 °C in IPL-41 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 2.6 g/L tryptose phosphate broth (TPB; Becton, Dickinson and Company, Franklin Lakes, NJ, USA), 10% (v/v) fetal bovine serum, and 0.05% (v/v) of an antibiotic and antimycotic mixture (penicillin, amphotericin B, and streptomycin; Sigma Aldrich, St. Louis, MO, USA), designated hereafter as complete IPL-41 medium.
The methods used for dry preservation and rehydration were based on the previous work (Watanabe et al. 2016), with the following modifications: Cells were incubated in a preconditioning medium (600 mM trehalose containing 10% (v/v) complete IPL-41 medium) for 48 h at 25 °C. The cells were then suspended in 400 µL of the preconditioning medium at a concentration of 1 × 108 cells/mL. Forty-microliter aliquots of this cell suspension were dispensed as droplets into 35-mm petri dishes. These dishes were immediately transferred into a desiccator and then maintained at < 10% relative humidity and 25 °C for 10 days. After rehydration by immersion in 1 mL of fresh complete IPL-41 medium, Pv11 cells were cultured and monitored until 14 days after rehydration.
Expression vectors
To isolate an effective and constitutive promoter in Pv11 cells, we previously cloned a 1624-bp genomic fragment (Δ0) comprising the 5′ upstream region of the glyceraldehyde 3-phosphate dehydrogenase (PvGapdh) gene (Okada et al. 2015); the PvGapdh gene was chosen, because it is highly and constitutively expressed in P. vanderplanki according to MidgeBase (http://bertone.nises-f.affrc.go.jp/midgebase/) (Gusev et al. 2014). Vectors containing a deletion series of the PvGapdh promoter (Fig. 1a) were constructed (Okada et al. 2015). Briefly, genomic DNA was isolated from Pv11 cells, and the 5′-upstream region of PvGapdh was amplified using a high-fidelity DNA polymerase, KOD-Plus- Neo (Toyobo, Osaka, Japan) with specific primer pairs. The amplified fragments obtained were inserted into the pIZ/V5-His vector (Thermo Fisher Scientific) to replace the original promoter regions (OpIE2 promoter) located upstream of the multi-cloning site (MCS) in the vector. At the same time, the zeocin resistance gene in the vector was replaced with the kanamycin/neomycin-resistance (Kan+/Neo+) gene. The vector that included the deletion variant of PvGapdh promoter (300–970) selected for further study and the Kan+/Neo+ gene was designated as pPGK (Supplementary Fig. 1A; Supplementary data 1). Subsequently, the remaining OpIE2 promoter in pPGK was replaced with the PvGapdh promoter (300–970), and the resulting vector was designated as pPGx2K (Supplementary Fig. 1B; Supplementary data 2).
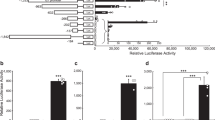
Exogenous gene expression system for Pv11 cells. a Schema representing the deletion clones for the PvGapdh promoter upstream of the AcGFP1 reporter gene. b Expression of AcGFP1 in Pv11 cells for each deletion clone was analyzed by fluorescence microscopy. All scale bars represent 100 µm. c Relative promoter activity of each deletion clone was measured by flow cytometry. Each plot represents the mean ± SEM of three samples
To construct vectors for the transient expression of the fluorescent proteins AcGFP1 and NLS (nuclear localization signal)-TagRFP, we isolated DNA fragments from pIRES2-AcGFP1 (TaKaRa Bio, Ohtsu, Japan) for the AcGFP1 gene, pTagRFP-N (Evrogen, Moscow, Russia) for the TagRFP gene, and pHsp70-Cas9 (Addgene, Cambridge, MA) for the NLS region. These DNA fragments were inserted into the MCS of the pPGK vector. The resultant vectors were named pPGK-AcGFP1 and pPGK-NLS-TagRFP.
To obtain stable expression of GFP, the AcGFP1 gene was inserted into the MCS of the pPGx2K vector; the resultant vector was named pPGx2K-AcGFP1.
Transfection of the expression vectors into Pv11 cells
For transient expression, expression vectors were transfected into Pv11 cells using the 4D-Nucleofector® system (Lonza, Basel, Switzerland) with SG Cell Line 4D-Nucleofector® X Kit S (Lonza) according to the manufacturer’s instructions. As the electroporation pulse code, DC134 was selected. Heterologous expression was allowed to proceed for 24 h (for flow cytometry) or 48 h (for microscopy). The fluorescence of expressed proteins was examined using a fluorescent microscope (Biozero BZ-X700; Keyence, Osaka, Japan) or flow cytometer (MoFlo Astrios, Beckman Coulter, Brea, CA, USA). Nuclei were detected by staining with Hoechst 33258 (Thermo Fisher Scientific).
Evaluation of promoter activity
Using the method of Ducrest et al. (2002), we evaluated the promoter activity of GFP reporter expression vectors by flow cytometry. A MoFlo Astrios cell-sorter equipped with 360-nm UV and 488-nm argon lasers was used to detect the fluorescence of propidium iodide (PI; a dead dell indicator) and AcGFP1, respectively. For the detection of PI and AcGFP1, red (670/40 nm) and green (405/30 nm) band pass filters, respectively, were used in combination with a 440-nm long pass filter. A minimum of 10,000 events in the fraction excluded dead cells was collected. To evaluate promoter activity, the mean GFP intensity was measured 2 days after AcGFP1-expressing cell cultures were separately transfected with the series of PvGapdh promoter-deletion series vectors.
Establishment of the GFP-expressing stable cell line (Pv11-KH cells)
The pPGx2K-AcGFP1 vector (10 µg) was transfected into Pv11cells (1 × 107 cells) by electroporation using an NEPA21 Super Electroporator (Nepa Gene, Chiba, Japan). The electroporation conditions were as follows: poring pulse (pulse voltage, 250 V; pulse width, 2 ms; pulse number, 6; voltage decay, 10%; voltage polarity, +) and transfer pulses (pulse voltage, 20 V; pulse width, 50 ms; pulse number, 5 for each polarity; voltage decay, 40%; voltage polarity, +/−). To select the GFP stable cell line, transfected cells were cultured in the complete IPL-41 medium with 400-µg/mL G418 (Thermo Fisher Scientific) for 1 month. Subsequently, GFP-expressing cells were collected with a MoFlo Astrios cell-sorter (Beckman Coulter) as shown in Supplementary Fig. 2. The isolated highly GFP-expressing cell line was designated as Pv11-KH and was used for further experiments.
Cell proliferation rate
To examine the proliferation rate of Pv11-KH cells after rehydration, the number of fluorescent GFP-expressing cells was counted using a Biozero BZ-X700 microscope (Keyence) with the BZ-H3C hybrid cell count software (Keyence).
Gene silencing by siRNA
The siRNAs for the AcGFP1 gene were designed using siDirect software (http://sidirect2.rnai.jp/design.cgi; Ui-Tei et al. 2004; Naito et al. 2009). The uniqueness of the candidate siRNAs was confirmed by BLASTN search (cutoff: 0.1) against the P. vanderplanki genome database MidgeBase (http://bertone.nises-f.affrc.go.jp/midgebase) to minimize off-target effects. All designed siRNAs were synthesized and annealed by Hokkaido System Science (Sapporo, Hokkaido, Japan). The sequences of the siRNAs are listed below.
-
AcGFP1-52-sens, 5′-GCUGAAUGGCGAUGUGAAUTT-3′ and
-
AcGFP1-52-anti, 5′-AUUCACAUCGCCAUUCAGCTT-3′ (for 52-siRNA);
-
AcGFP1-443-sens, 5′-CACAAUGUGUACAUCAUGATT-3′ and
-
AcGFP1-443-anti, 5′-UCAUGAUGUACACAUUGUGTT-3′ (for 443-siRNA);
-
AcGFP1-480-sens, 5′-GCAUCAAGGUGAACUUCAATT-3′ and
-
AcGFP1-480-anti, 5′-UUGAAGUUCACCUUGAUGCTT-3′ (for 480-siRNA).
Simultaneously, we designed 2 types of siRNA with randomized sequences as negative controls. BLASTN searches (cutoff: 0.1) showed that these sequences were not present in the genome of P. vanderplanki. The sequences of these siRNAs are listed below.
-
NC1-sens, 5′-GCACUGCUACGAUCGUUAATT-3′ and
-
NC1-anti, 5′-UUAACGAUCGUAGCAGUGCTT-3′ (for NC1);
-
NC2-sens, 5′-GUAGAGAGCGCGAUCUAUATT-3′ and
-
NC2-anti, 5′-UAUAGAUCGCGCUCUCUACTT-3′ (for NC2).
siRNA was transfected into stable GFP-expressing Pv11-KH cells by electroporation. Briefly, Pv11 cells were transfected with either siRNAs (0.5-μM final concentration) or TE [10 mM Tris–HCl (pH 8) and 1-mM EDTA] buffer as a mock treatment using the 4D-nucleofector (Lonza). Three days after transfection, the fluorescence of the transfected cells was evaluated with a Biozero BZ-X700 microscope (Keyence) and a MoFlo Astrios cell-sorter (Beckman Coulter). Concomitantly, the expression of AcGFP1 in the transfected cells was examined by the western blot analysis.
Western blot analysis
For SDS-PAGE, cells were collected by centrifugation (700×g for 3 min) and lysed in RIPA buffer (Nacalai Tesque, Kyoto, Japan) for 1 h at 4 °C. The concentrations of the obtained crude lysates were quantified with a Qubit protein assay kit (Thermo Fisher Scientific). The lysates (7.5 µg/lane) were applied to SDS-PAGE and the separated proteins were transferred to a polyvinylidene fluoride transfer membrane using the Trans-blot Turbo system (Bio-Rad, Hercules, CA, USA). Immunoblotting analysis was performed with an SNAP i.d. system (Millipore, Billerica, MA, USA). The blot was treated with anti-AcGFP1 antibody (Living Colors Full-Length A.v. Polyclonal Antibody; Clontech, Mountain View, CA, USA; 1:1000) and subsequently with peroxidase-labeled goat anti-rabbit IgG (Thermo Fisher Scientific). The immunoreaction signal was detected with ECL Plus Western Blotting Detection Reagents (GE Healthcare, Little Chalfont, UK) and visualized with a luminescent image analyzer (LAS-3000 mini; Fujifilm, Tokyo, Japan). After detection of bands corresponding to immunoreactive proteins, the blotted membrane was stained with CBB stain (Bio-safe Coomassie Stain; Bio-Rad) to validate the experiment (Supplementary Fig. 3). Band intensities were quantified with the ImageJ 1.51e program (https://imagej.nih.gov/ij/index.html).
Results and discussion
Expression of exogenous protein in Pv11 cells
The first step in developing an exogenous gene expression system for Pv11 cells was to isolate a strong constitutive promoter in P. vanderplanki. From transcriptome data in the P. vanderplanki genome database MidgeBase (Gusev et al. 2014), we previously selected the glyceraldehyde-3-phosphate dehydrogenase (PvGapdh) gene as constitutively expressed at high levels throughout desiccation and rehydration processes (Okada et al. 2015). We cloned 1.6 kb of the 5′-upstream region of PvGapdh, and we compared GFP fluorescence in cells transfected with deletion mutants of this putative promoter using a fluorescent microscope (Okada et al. 2015). Here, we accurately evaluated the promoter activity of the deletion clones. Once the GFP expression vector containing the whole 1.6 kb-fragment of PvGapdh promoter (Δ0) was constructed, we made deletion mutant series of the promoter in the vector (Fig. 1a). We then transfected each constructed vector into Pv11 cells, confirmed the expression of GFP using a fluorescence microscope (Fig. 1b), and measured the promoter activity of each promoter candidate with a flow cytometer (Fig. 1c). Comparison of promoter activity among the deletion mutants showed that GFP expression in the 300–970 vector was only slightly weaker than that in the Δ0 or Δ300 vectors (Fig. 1c). Interestingly, even though the Δ770 vector contains well-known minimal promoter motifs, such as CAAT-box and TATA-box, this vector exhibited almost no promoter activity in the cells. This result indicates that some unknown strong enhancer or promoter might occur in the region from 300 to 770 relative to the 5′ end of the 1.6-kb 5′-upstream region. Because use of the shortest effective promoter is best when constructing an expression vector, to avoid size limitations when inserting the gene of interest, the PvGapdh promoter-deletion mutant 300–970 (Fig. 1a) was selected as containing the most suitable promoter for the construction of a new constitutive expression system in Pv11 cells. The new vector including this engineered promoter was designated as pPGK vector (Supplementary Fig. 1A).
Signal peptides play a key role in protein targeting in the cell. In particular, the NLS is important to artificially deliver expressed exogenous proteins to the nucleus. For example, exogenous proteins, such as transcription factors, must be localized in the nucleus to function. Hence, to investigate whether a typical commercially available NLS derived from SV40 large T-antigen was functional in Pv11 cells, we transfected a construct encoding NLS-TagRFP fusion protein into Pv11 cells. Using a cell-permeant nuclear counterstain reagent, Hoechst 33342, we demonstrated that the fluorescence of NLS-TagRFP was localized in the nucleus (Fig. 2). This result indicates that the commercially available NLS could function in Pv11 cells and opens the way to future genome editing using the CRISPR/Cas system.
Subcellular localization of NLS-TagRFP fusion protein in Pv11 cells. a Red spots show the fluorescence of the NLS-TagRFP fusion protein. b Fluorescence of Hoechst 33342 nucleic acid stain reagent shows the locations of the nuclei in Pv11 cells. c Bright filed image of Pv11 cells. d Arrowheads in the merged image indicate overlapping spots of fluorescence by NLS-TagRFP and Hoechst33342
Thus, our newly developed vector system allows effective expression of exogenous proteins in Pv11 cells.
Establishment of a GFP-expressing stable cell line of Pv11
Once the techniques for efficient gain-of-function were established, we then explored whether exogenous GFP-expressing Pv11 cells could be used as a tool to examine desiccation tolerance. Hence, we established a GFP-expressing stable Pv11 cell line, designated as Pv11-KH, which was selected by flow cytometry and isolated by cell sorting (Supplementary Fig. 2), and we assessed whether this cell line possessed a similar desiccation tolerance to that of the Pv11 cell line. When Pv11-KH cells were desiccated for 10 days and then rehydrated for 1 day, 6.30 ± 0.51% of the cells were GFP-expressing following the rehydration step; this proportion is slightly less than the survival rate of original Pv11 cells as previously observed (Watanabe et al. 2016). Therefore, we consider that the GFP-expressing cells observed after rehydration were surviving cells retained fluorescence. Nearly, all Pv11-KH cells showed GFP fluorescence throughout the process of desiccation, but after rehydration, the number of GFP-expressing cells dropped drastically. The disappearance of GFP fluorescence upon rehydration is likely due to temporal membrane leakage and GFP diffusion into the medium. During culture after rehydration, the number of GFP-positive cells continuously increased (Fig. 3), indicating successful proliferation of the dry-preserved Pv11-KH cells. Even more, after passages, all progeny of the rehydrated Pv11-KH cells retained fluorescence. These findings suggest that the Pv11-KH cells retained a desiccation tolerance similar to that of original Pv11 cells.
Efficient gene silencing by siRNA in Pv11 cells
Our exogenous expression system was established as a gain-of-function technique for Pv11 cells; however, loss-of-function is also needed as an important tool in terms of reverse genetics. To validate the efficiency of RNA interference (RNAi) for Pv11 cells, we performed gene knockdown by siRNA, targeting AcGFP1 in Pv11-KH cells. The siRNAs were transfected into Pv11-KH cells, and 3 days later, the GFP fluorescence was assessed by fluorescence microscopy (Fig. 4a) and flow cytometry (Fig. 4b). The fluorescence was significantly suppressed by transfection with each GFP-specific siRNA, but not the NC1 and NC2 control siRNAs. To confirm gene silencing, crude extracts from the cells were subjected to the western blotting analysis. Transfection with GFP-specific siRNAs suppressed the accumulation of AcGFP1 protein to approximately 7.5–34.6% of that in the non-siRNA-transfected Pv11-KH cells (Fig. 4c). These results suggest that gene silencing by siRNA is effective in Pv11 cells.
Gene silencing of GFP expression in Pv11-KH cells. a Fluorescence images indicate AcGFP1 expression in Pv11-KH cells transfected with each AcGFP1-specific siRNA (52-siRNA, 422-siRNA, 480-siRNA), with control random siRNA (NC1, NC2), or with TE buffer only (Mock), 3 days after transfection. All scale bars represent 100 µm. b Histogram showing the relative GFP fluorescence of Pv11-KH cells transfected with each siRNA. c Accumulation of AcGFP1 protein in siRNA-transfected cells was evaluated by the western blot analysis. Relative intensities of the band for AcGFP1 compared with mock-transfected cells are indicated below each lane
Conclusion and perspectives
Anhydrobiosis is a peculiar biological phenomenon. Thus far, there has been no model system to investigate the molecular mechanisms underlying anhydrobiosis in animal cultured cells; therefore, most of these mechanisms still need to be elucidated. Here, we developed both gain- and loss-of-function techniques for the anhydrobiotic cultured cell line Pv11. Currently, we are optimizing genome-editing systems, such as CRISPR/Cas and TALEN, for Pv11 cells. We believe that the application of Pv11 cells as a new model system, together with a combination of these newly developed techniques, will help unveil the organization of the gene network involved in the mechanism of extreme desiccation tolerance.
References
Cornette R, Kanamori Y, Watanabe M et al (2010) Identification of anhydrobiosis-related genes from an expressed sequence tag database in the cryptobiotic midge Polypedilum vanderplanki (Diptera; Chironomidae). J Biol Chem 285:35889–35899. doi:10.1074/jbc.M110.150623
Ducrest AL, Amacker M, Lingner J, Nabholz M (2002) Detection of promoter activity by flow cytometric analysis of GFP reporter expression. Nucleic Acids Res 30:e65. doi:10.1093/nar/gnf064
Erkut C, Penkov S, Khesbak H et al (2011) Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol 21:1331–1336. doi:10.1016/j.cub.2011.06.064
Gusev O, Suetsugu Y, Cornette R et al (2014) Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat Commun 5:4784. doi:10.1038/ncomms5784
Hatanaka R, Hagiwara-Komoda Y, Furuki T et al (2013) An abundant LEA protein in the anhydrobiotic midge, PvLEA4, acts as a molecular shield by limiting growth of aggregating protein particles. Insect Biochem Mol Biol 43:1055–1067. doi:10.1016/j.ibmb.2013.08.004
Kikawada T, Nakanahara Y, Kanamori Y et al (2006) Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun 348:56–61. doi:10.1016/j.bbrc.2006.07.003
Li S, Chakraborty N, Borcar A, Menze MA, Toner M, Hand SC (2012) Late embryogenesis abundant proteins protect human hepatoma cells during acute desiccation. Proc Natl Acad Sci USA 109:20859–20864. doi:10.1073/pnas.1214893109
Naito Y, Yoshimura J, Morishita S, Ui-Tei K (2009) siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform 10:392. doi:10.1186/1471-2105-10-392
Nakahara Y, Imanishi S, Mitsumasu K et al (2010) Cells from an anhydrobiotic chironomid survive almost complete desiccation. Cryobiology 60:138–146. doi:10.1016/j.cryobiol.2009.10.004
Okada J, Kikuta S, Gusev O et al (2015) Construction of optimized CRISPR/Cas system to reveal the mechanisms of anhydrobiosis in the Sleeping Chironomid. Cryobio Cryotech 61:69–73
Sakurai M, Furuki T, Akao K et al (2008) Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilum vanderplanki. PNAS 105:5093–5098. doi:10.1073/pnas.0706197105
Shimizu T, Kanamori Y, Furuki T et al (2010) Dessication-induced structuralization and glass formation of group 3 Late Embryogenesis Abundant protein model peptides. Biochemistry 49:1093–1104. doi:10.1021/bi901745f
Ui-Tei K, Naito Y, Takahashi F et al (2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32:936–948. doi:10.1093/nar/gkh247
Watanabe M (2006) Anhydrobiosis in invertebrates. Appl Entomol Zool 41:15–31
Watanabe M, Kikawada T, Okuda T (2003) Increase of internal ion concentration triggers trehalose synthesis associated with cryptobiosis in larvae of Polypedilum vanderplanki. J Exp Biol 206:2281–2286
Watanabe K, Imanishi S, Akiduki G, Cornette R, Okuda T (2016) Air-dried cells from the anhydrobiotic insect, Polypedilum vanderplanki, can survive long term preservation at room temperature and retain proliferation potential after rehydration. Cryobiology 73:93–98. doi:10.1016/j.cryobiol.2016.05.006
Acknowledgements
We are grateful to R. Hatanaka and T. Furusawa for helping perform plasmid construction and cell-sorting, respectively. We also thank T. Shirotori and Y. Kikuzato for technical assistance. This work was supported in part by the Grants-in-Aid from Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant Numbers 16K18827, 16J09151, 16K07308, 15H02378, 25252060, and 16K15073); research fellowship of JSPS for Young Scientists (#13J08784); a Grant for Basic Science Research Projects from the Sumitomo Foundation (140890); a Grant for the Narishige Zoological Science Award 2016; construction of siRNA and plasmids were supported by Russian Science Foundation grant for international groups (No. 14-44-00022).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by H. Atomi.
This article is part of a special feature based on the 11th International Congress on Extremophiles held in Kyoto, Japan, September 12–16, 2016.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sogame, Y., Okada, J., Kikuta, S. et al. Establishment of gene transfer and gene silencing methods in a desiccation-tolerant cell line, Pv11. Extremophiles 21, 65–72 (2017). https://doi.org/10.1007/s00792-016-0880-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-016-0880-4