Abstract
Objectives
Gingival crevicular fluid (GCF), the inflammatory infiltrate within the crevicular sulcus, is of great importance for diverse processes in the oral cavity and has a high impact in oral sciences. It is assumed to serve as a source of biomarkers for systemic or periodontal diseases and mediators of orthodontic tooth movement. In order to characterize the protein content of the GCF in an unbiased and complete approach, we employed mass spectrometry (MS), which allows not only the identification, but also the quantification of these proteins. In samples obtained from patients suffering from periodontitis, this method is often limited due to the presence of highly abundant serum albumin deriving from serum. The aim of this investigation was to employ a protein precipitation procedure for the efficient depletion of serum albumin from GCF samples.
Materials and methods
GFC samples collected from five adult periodontitis patients were fractionated by trichloroacetic acid/acetone precipitation and the resulting soluble and pelleted fractions were analyzed by SDS-PAGE and high-resolution mass spectrometry.
Results
Trichloroacetic acid/acetone precipitation was successfully employed as a protein precipitation procedure for the efficient depletion of serum albumin from GCF samples. Careful analysis revealed that the precipitation step reduced the serum albumin content efficiently, and increased subsequent protein identifications by 32%. Three hundred seventeen proteins could only be identified with this new approach.
Conclusion
The increased coverage of the GCF proteome will help improve our understanding of molecular mechanisms in the periodontium during pathogenesis of periodontitis.
Clinical relevance
Our new albumin depletion strategy combined with high-resolution mass spectrometry can be used to effectively monitor the molecular signals of the periodontium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The periodontium protects the tooth from the bacteria in the oral cavity. To supply protection from infections, the periodontium secretes an inflammatory infiltrate within the crevicular sulcus even in situations of clinical health [1,2,3]. This body fluid, the gingival crevicular fluid (GCF), contains a mixture of biomolecules derived from serum, host cells and oral bacteria [4, 5]. Compared to saliva, GCF has greater potential of containing periodontal or systemic biomarkers because of its site-specific nature [1]. The major limitation for the study of this complex fluid is the availability of very small quantities per site, approximately 0.2–0.5 μl [2]. Upon inflammation of the periodontium, the alveolar bone and the surrounding tissues are damaged, and GCF formation becomes exudative [6]. Under these conditions, the proportion of serum proteins in GCF increases from approx. 30 to 70% [7].
The investigation of GCF is of great interest for almost all dental disciplines. Mass spectrometry-based proteome analysis has already been demonstrated to be a valuable tool in periodontal research. The analysis of GCF samples during orthodontic tooth movement (OTM) helps understanding this movement substantially [8, 9]. Mass spectrometry (MS)-based proteomics does not only help characterize host-related proteins though. The field of metaproteomics describes the large-scale characterization of entire protein complements including both the host and the environmental microbiota [10]. Protein changes of biofilms in response to the host tissue have recently been identified by mass spectrometry [11]. Using mass spectrometry for the investigation of proteins originating from bacteria in GCF opens a large field for periodontal research.
Previous studies for the investigation of protein expression patterns in GCF have used targeted approaches such as Western blotting or enzyme-linked immunosorbent assays (ELISA) [12, 13]. Compared to these techniques, unbiased approaches such as mass spectrometry-based proteome analysis have great advantages. They have been successfully used to discover biomarkers for the detection of various diseases [14]. In recent years, the development of new mass spectrometers and sample preparation strategies have enabled substantial progress resulting in higher protein identification and quantification rates [15]. Mass spectrometric analysis of proteins is commonly based on the ionization and subsequent fragmentation of surrogate peptides generated by enzymatic proteolysis, and the high-resolution analysis of both their intact and fragmented mass-to-charge (m/z) ratios. This allows for the determination of molecular weight, sequence and, and with novel technologies, also structure of peptides and their associated proteins [16]. Relative quantification, i.e., the determination of up- or downregulation between two or more samples (e.g., health and disease) is instructive for studying the pathology of a disease, and may be achieved, e.g., through the use of isotopic labeling strategies.
In the analysis of body fluids, however, a limitation is frequently imposed by the presence of high abundance proteins (e.g., albumin in serum or GCF) that superimpose on low abundant proteins, which typically include many biomarkers, and prevent their reliable identification and quantitation. In recent years, MS technology has been used for determining GCF protein content [3, 17,18,19]. Most authors reported limitations due to the presence of highly abundant serum-derived proteins especially in periodontally diseased sites [7]. Our aim in this study was therefore to find a method to deplete serum albumin in GCF, and thus to enable the detection of otherwise undetectable protein components in GCF by mass spectrometry.
Material and methods
This study was approved by the responsible ethics committee (University of Goettingen) with protocol number 23/7/15. Informed written consent was obtained from all patients. GCF was collected from periodontally inflamed sites from a total of five subjects. Subjects had at least 20 teeth and had been diagnosed with chronic periodontitis (as per the criteria of the International Workshop for a Classification of Periodontal Diseases and Conditions from 1999)) [20]. The distance from cementoenamel junction to the base of the crevice (CAL) was measured and patients having at least four teeth with a CAL of 5 mm or more (in at least two different quadrants) were selected. The preselected specific inflamed sites were defined by pocket depth of 5–7 mm. Exclusion criteria were the use of premedications or antibiotics in the past 6 months, systemic conditions which may affect the progression of periodontitis, pregnancy or lactation, alcohol or drug abuse. Differences between genders and ages were not analyzed since the study was not designed or aimed to define such.
Collection of GCF
The collection of GCF was performed according to Carneiro et al. 2014. GCF was collected from up to six sites per subject at the lower frontal incisors and canines using PerioPaper® Gingival Fluid Collection Strips (ORAFLOW, Smithtown, NY, USA). All volunteers were asked not to eat, drink, brush their teeth or to use any type of mouthwash 2 h prior to fluid collection. Before the collection, the selected sites were cleaned with polishing cups (Produits Dentaire SA, Vevey, Switzerland) without any form of polishing paste and subjected to thorough washing with the dental unit’s air-water syringe. The selected sites were isolated from salivary contamination with cotton rolls, and air-dried afterwards. A sterile PerioPaper® strip was gently inserted into the sulcus and left in place for 30 s. Strips contaminated with blood were discarded and mechanical irritation was avoided. The PerioPaper® strips were placed into Eppendorf tubes (Eppendorf, Hamburg, Germany) and kept frozen at − 80 °C until further processing. Elution of GCF proteins from PerioPapers® was performed using 30 μl ammonium bicarbonate solution (NH4HCO3) 50 mM (pH 8.0). The tube was vortexed in a Thermomix (Eppendorf, Hamburg, Germany) for 10 min at room temperature and 850 rpm. Thirty microliters of the protein-NH4HCO3 eluate was collected using Pierce Spin Columns (Thermo Fisher, Bremen, Germany). The eluted GCF proteins were aliquoted into two portions of 15 μl each, frozen and stored at − 80 °C until further use.
Sample processing
In order to deplete highly abundant serum albumin in GCF, a trichloroacetic acid/acetone precipitation protocol was used for one aliquot per subject. Chen et al. described a modified protein precipitation procedure for the efficient removal of albumin from serum [21]. We modified this protocol in the following way: 60 μl of 10% (v/v) ice-cold TCA-acetone solution were added to the sample (GCF solved in NH4HCO3), the solution vortexed and incubated for 2 h at − 20 °C. After centrifugation for 30 min at 13000 rpm/4 °C, supernatant I was removed and further processed, the precipitate I was collected, 0.5 ml ice-cold acetone (100%) was added to the supernatant and incubated on ice for 15 min. After centrifugation for 30 min at 13000 rpm at 4 °C, supernatant II was removed and further processed, while precipitate II was collected. Supernatant II was lyophilized with a Speedvac concentrator (Centrifuge 5810, Eppendorf, Hamburg, Germany) resulting in precipitate III (Fig. 1).
Experimental procedure: collection and elution of GCF. The eluted GCF was aliquoted in two equal portions (named samples A and B). A was analyzed without prior condition. For B, we performed precipitation with TCA-acetone 10% v/v. To test which proteins cannot be identified by mass spectrometry due to prior precipitation of GCF with TCA-acetone (precipitate I), the supernatant of this precipitation step was again precipitated with 100% acetone (precipitate II). The supernatant of this second precipitation step was dried in a SpeedVac (Precipitate III). SDS-PAGE was performed followed by in-gel trypsin digestion and protein identification by mass spectrometry
One-dimensional polyacrylamide gel electrophoresis (SDS-PAGE)
For SDS-PAGE, the NuPAGE® system was used according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). The gel was stained overnight with colloidal coomassie G-250 [22]. Aqueous buffers were sterile filtered (Millipore filter system, Billerica, USA).
In-gel trypsin digestion and extraction of proteins
Using an in-house manufactured gel cutter, each lane was cut into 13 equidistant slices. After further dicing, samples were subsequently reduced (100 mM Cleland’s reagent, Calbiochem, Darmstadt, Germany, in 0.1 M ammonium carbonate, pH 8.0), alkylated, digested with trypsin, and peptides were extracted as described [23]. All incubation steps were performed with Thermomix (Eppendorf, Hamburg, Germany) at 1050 rpm for 15 min/20 °C, unless otherwise stated. Gel dices were washed with 150 μl water for 5 min and subsequently dehydrated with 150 μl acetonitrile (ACN). The gel dices were dried for 5 min, then rehydrated with 100 μl reducing solution, subsequently incubated at 56 °C for 50 min, and dehydrated with 150 μl ACN. In a next step, the proteins were alkylated with 100 μl 60 mM Iodacetamide (Sigma-Aldrich, Steinheim, Germany) in 0.1 M ammonium carbonate for 20 min at 26 °C in the dark. The gel pieces were washed with 150 μl ammonium carbonate (pH 8.0) adding 150 μl ACN. Then the solution was removed and the gel dices were dehydrated with 150 μl ACN and dried in a hood for 10 min. The gel dices were now subjected to in-gel digestion by incubation for 15 min with 10 μl of digestion buffer containing 45 μl trypsin (12.5 μg/ml, Roche, Basel, Switzerland), 150 μl 0.1 M ammonium carbonate (pH 8.0), 15 μl CaCl2 (5 mM) and 150 μl H2O, and stored on ice. After 15 min, the gel dice were again covered with 5–10 μl of digestion buffer and kept on ice for 20 min. If the dices absorbed to much digestion buffer, they were covered with buffer without trypsin [150 μl 0.1 M ammonium carbonate (pH 8.0), 15 μl CaCl2 (5 mM) and 195 μl H2O]. Trypsin digestion was carried out at 37 °C overnight.
Mass spectrometry
Protein digests were analyzed on a nanoflow chromatography system (Eksigent nanoLC425) hyphenated to a hybrid triple quadrupole-time of flight mass spectrometer (TripleTOF 5600+) equipped with a Nanospray III ion source (Ionspray Voltage 2200 V, Interface Heater Temperature 150 °C, Sheath Gas Setting 10) and controlled by Analyst TF 1.6 software build 6211 (all equipment Sciex, Darmstadt, Germany). In brief, samples were dissolved in 20 μL loading buffer (2% aqueous acetonitrile vs. 0.1% formic acid). For each analysis, 25% of each gel slice sample were concentrated and desalted on a trap column (0.15 mm ID × 20 mm, Reprosil-Pur120 C18-AQ 5 μm, Dr. Maisch, Ammerbuch-Entringen, Germany, 60 μl loading buffer) and separated by reversed phase-C18 nanoflow chromatography (0.075 mm ID × 150 mm, Reprosil-Pur 120 C18-AQ, 3 μm, Dr. Maisch, linear gradient 30 min 5% > 35% acetonitrile vs. 0.1% formic acid, 300 nL/min, 50 °C).
Qualitative LC/MS/MS analysis was performed using a Top15 data-dependent acquisition method with an MS survey scan of m/z 380–1250 accumulated for 250 ms at a resolution of 35.000 FWHM. MS/MS scans of m/z 180–1600 were accumulated for 100 ms at a resolution of 17.500 FWHM and a precursor isolation width of 0.7 FWHM, resulting in a total cycle time of 1.8 s. Precursors above a threshold MS intensity of 150 cps with charge states 2+, 3+ and 4+ were selected for MS/MS, the dynamic exclusion time was set to 15 s. Two technical replicates per sample were acquired.
Data analysis
Protein identification was achieved using ProteinPilot software (v5.0.0.0, SCIEX, Darmstadt, Germany). Proteins were identified at “thorough” settings against the UniProtKB human reference proteome v2014.10 (64,336 protein entries) along with a set of 51 contaminants commonly identified in our laboratory. Scaffold Software (v4.4.8, Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Paragon algorithm [24]. Protein identifications were accepted if they could be established at greater than 33.0% probability to achieve an FDR less than 1.0% and contained at least two identified peptides. Protein probabilities were assigned by the protein Prophet algorithm [25]. Proteins which contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Results
The aim of this study was to develop an approach for unbiased, quantitative protein analysis from gingival crevicular fluid (GCF) by mass spectrometry in the presence of high abundance serum-derived proteins. Previous studies encountered limitations due to the presence of high abundant serum-derived proteins especially in periodontally diseased sites [1, 7]. We applied TCA-acetone precipitation to deplete GCF of serum albumin, and found this approach efficient for increasing protein identifications in GCF. To validate the method, we followed the experimental procedure outlined in Fig. 1. Eluted GCF of individual subjects was aliquoted into two equal samples, A and B. Only sample B was further processed by sequential precipitation of proteins with TCA-acetone to yield precipitates I, II, and III (see Material and methods section). Precipitate I contained a wide spectrum of small to very large proteins. To test which proteins escaped their identification by mass spectrometry due to the precipitation step with TCA-acetone, the supernatant of precipitation I was again precipitated with 100% acetone (precipitate II). Precipitate II contained a dominant band representing serum albumin, and in addition other, mostly smaller proteins. Thus, the purpose of removing albumin from precipitate I seemed to be largely fulfilled. The supernatant of the second precipitation was dried in a SpeedVac to yield precipitate III, which did not contain significant amounts of protein. With this experimental setup, all proteins of sample B were visualized by gel electrophoresis (Fig. 1). Our analysis demonstrated that the precipitation procedure presents an efficient method for proteome analysis from GCF samples. For all five patients, we examined the influence of our procedure on the rates of protein identification by mass spectrometry of sample A (GCF without precipitation) and sample B/precipitates I–III (GCF with prior TCA-acetone precipitation), respectively, using two technical replicates (Fig. 2 and Supplementary Table 1).
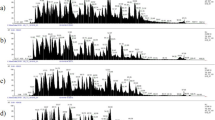
SDS-PAGE of GCF samples from five patients. The sample was aliquoted in two equal portions A and B. Lane 1: molecular weight marker. Lane 2: unprecipitated GCF portion A. Lanes 3–5: precipitation steps of the portion B. In all five samples, the serum albumine protein band shifts from precipitate I to precipitate II. In precipitate III, there is no remaining protein detectable with coomassie staining
The concentration of individual proteins in a sample can be profiled using spectral counting, where the number of successful MS/MS spectral assignments to the sequence of a protein correlates to its concentration [26]. As expected, most protein counts in sample A were scored for serum albumin. By precipitation with TCA/acetone, the albumin concentration is reduced by about 84% in PI (average of five patients). The majority of the precipitated albumin is found in PII, while relatively few total spectrum counts were found for serum albumin in PIII (Table 1). Taken together, the results from gel electrophoresis and mass spectrometry demonstrate that precipitation with TCA-acetone represents an efficient method to reduce serum albumin and to increase protein identifications in GCF by 32% (pool of five subjects).
In sample A, 990 proteins or protein clusters could be identified, while 1305 proteins were found in sample B/precipitates I–III (Fig. 3a). Nine hundred eighty-eight proteins were identified in both in samples A and B, with only two proteins identified solely without fractionation (Table 2). On the contrary, 317 proteins could only be identified with the new approach.
Pool of GCF samples from five patients. a In sample A (GCF without prior precipitation), 990 proteins were identified. With the new methodical preparation procedure of sample B, this number could be increased to 1305 identifications (PI–PII) while only two proteins could not be identified due to sample preparation with TCA/acetone. b The comparison of sample A and precipitate I of sample B shows an increase of 201 proteins due to albumin depletion with TCA/acetone. c Despite the higher abundance of albumin, PII contains nearly as much proteins as PI while PIII contains only one protein that is not identified in either PI or PII
For future use in GCF proteome studies, it was important to see whether it is necessary to analyze all precipitates I, II and III, or if it is sufficient to analyze precipitate I only (Fig. 3c). As already suggested by gel electrophoresis, it turned out that analysis of PIII shows no additional benefit, as only one protein identification is covered by PIII (Table 3). Analysis of PII identified 113 proteins which are not already identified in PI. We also compared PI directly to the unfractionated sample A, to figure out if analysis of PI only would already provide sufficient benefit for routine proteome analysis. In this comparison, 37 proteins are identified exclusively in sample A without fractionation, and 238 proteins exclusively in PI (Fig. 3b). Another important aspect of sample fractionation in proteome analysis is whether it will conserve or improve information not only about protein identity, but also about protein quantity. To evaluate the performance of our separation strategy in this regard, we correlated the averaged spectral count (SC) values of proteins detected in the unfractionated samples A with those in precipitates PI and PII (Fig. 4). SC values in PI showed a strong linear correlation with those in samples A (Pearson 0.9325), with the slope of 1.16 confirming overall enrichment. Values in PII, however, correlated poorly (Pearson 0.7899), indicating that PII does not retain information about relative protein quantities in the unprecipitated sample. We therefore chose to proceed with PI as the relevant fraction. Table 4 displays a selection of 25 potentially important proteins identified only in PI of sample B. The entire proteome of PI contains 1191 proteins. Three hundred eighty-two of these proteins are identified in all samples. One hundred sixty-two proteins are identified in four of five subjects and 169 proteins are identified in three of five subjects (supplementary Table 2). We defined these 707 proteins identified in PI in at least three subjects as the “GCF core proteome” and interrogated these globally for their relevance in the context of oral biology.
a Spectral counts correlation plot PI vs. unprecipitated sample A (577 shared protein clusters excluding human serum albumin). Slope > 1 and the Pearson correlation coefficient indicate that quantitative information is well retained. b Spectral counts correlation plot PII vs. unprecipitated sample A (521 shared protein clusters excluding human serum albumin). Slope and the Pearson correlation coefficient indicate that quantitative information deteriorates in the serum albumin co-precipitate
We performed a functional enrichment analysis of the GCF core proteome using the DAVID Functional Annotation tool [27, 28] and interrogated it for enrichment of gene ontology (GO) terms (Table 5). Cellular component (CC) enrichment in GO terms showed that 81.9% of detected proteins are associated with extracellular exosomes, an observation that was previously made in an in-depth unbiased proteome study in GCF [1]. Of note is an overrepresentation of proteins associated with either the extracellular matrix (ECM), focal adhesion or cell-cell adherens junctions, highlighting the role of mechanical stabilization in the gingival crevice. Enrichment analysis for biological process (BP) terms additional points to important immune response-related functional circuits such as complement activation. In order to check the relevance of the identified proteins for oral biology, we annotated the list of GCF proteins according to their biological significance and molecular function using the Scaffold software (Table 5). For example, the group of proteins with chemoattractant function can be highly interesting for research on periodontitis. Table 4 exemplary represents the four proteins with chemoattractant functions in the proteome of GCF from a patient suffering from periodontitis (Table 5). All in all, global functional enrichment analysis of the identified proteins’ annotations in PI is in very good agreement with existing knowledge about the protein complement of the GCF.
Discussion
The periodontium is one of the most important structures in the oral cavity and responsible for numerous functions of the human body. Many questions regarding the molecular biology of this important structure are not yet answered. Investigating the entire protein complement of GCF is therefore of great importance for dental research. Although GCF can be obtained in a simple, non-invasive manner, its analysis offers insights that currently can only be obtained by much more elaborate approaches such as X-ray diagnostics [1]. In our opinion, mass spectrometry-based proteome analysis is the most promising approach to create a molecular base for investigations of GCF.
In our study, we show that one of the major problems in the investigation of small volumes of gingival crevicular fluid by mass spectrometry, i.e., the presence of high abundance serum-derived albumin, can be mitigated by simple methodology. The use of TCA/acetone precipitation for depletion of serum albumin was described for blood/serum samples [21]. Earlier results indicated that human serum albumin is soluble in organic solvents such as methanol, ethanol or acetone after precipitation with TCA [29, 30]. We show that this method provides great benefit for the mass spectrometric analysis of GCF collected from periodontitis patients. To validate our observations, we used an experimental strategy, which visualized not only the precipitate, but also the protein content of the supernatant. Thus, we could confirm that it is sufficient to analyze only precipitate I by mass spectrometry, and still increase identification rates substantially. The precipitation method required minimally higher preparation efforts, but importantly did not increase the required mass spectrometry instrument time. The identification rate could again be slightly increased by additional mass spectrometric analysis of precipitate II (in this case 113 proteins). For particular issues, this may be of importance, but it leads to a doubling of measurement time with a relatively small increase of the number of identified proteins.
Our investigation shows that MS of precipitate III can be omitted because no additional proteins could be identified. The total spectrum counts of serum albumin (Table 1) confirm the efficiency of this sample preparation procedure. As expected, the majority of serum albumin is found in precipitate II. Albumin acts as a carrier-protein and transports biomolecules within the blood and therefore is likely to bind proteins of interest such as cytokines and chemokines. Hence the depletion of albumin may be a methodological problem when its removal also leads to depletion of such important proteins. Our results show that with TCA/acetone precipitation prior to MS analysis of GCF, only two proteins were no longer found due to the pretreatment. None of these proteins has been previously discussed in the context of oral medicine (Table 2). On the other hand, in precipitate I, 316 proteins were detected in addition to the overlap with proteins identified in sample A (Fig. 3). Many of these proteins are of great importance for oral medicine. A selection of these proteins is annotated in Table 4, and briefly discussed below to show the convenience of the new sample precipitation procedure. In periodontal research, there is increasing evidence that. azurocidin serves as a potential biomarker for periodontitis [31]. Cathepsin B correlates positively with clinical parameters of disease severity in untreated chronic periodontitis [32]. New studies suggest that mitogen-activated protein kinases play a significant part of the ontogeny of inflammation in periodontal disease resulting in alveolar bone loss [33]. Earlier results show an association of coronin 1A with greater pocket depths and signs of inflammation [19]. MMP-9 is involved in different phases of collagen remodeling and in degradation of the PDL extracellular matrix during orthodontic tooth movement [34]. MS technology also allows the identification of novel uncharacterized proteins like uncharacterized protein B4E1Z4 and protein A0A0J9YY99 in body fluids. Noteworthy, all proteins discussed above would not have been identified without TCA/acetone precipitation prior to MS analysis.
When using our albumin depletion method, immunoglobulin proteins become the more abundant components in precipitate I (supplementary Table 1). For certain proteomic research of GCF, it may be desirable to remove immunoglobulins and albumin from the samples. In the context of periodontitis, however, immunoglobulins are of special interest and should not be depleted. Our study covers a relatively small number of subjects. However, since the method is sufficiently investigated for the MS analysis of serum [21], we do not think that an increased number of cases is necessary to confirm the utility of our sample preparation method.
The application of MS technology has proven extremely suitable for the identification of proteins in GCF with respect to specificity and sensitivity [1,2,3]. Further improvements require special attention, but are nevertheless conceivable. The major difficulty was not so much the extremely small amount of GCF, since the sensitivity of modern mass spectrometers more than compensates for the low protein amounts contained, but rather the overwhelming amount of especially albumin from contaminating blood. This is a common challenge not only for the analysis of GCF, but also several other body fluids. By including a TCA precipitation step before MS, we introduce here a simple and efficient procedure to improve the MS detection of proteins for the investigation of GCF.
References
Rody WJ Jr, Holliday LS, McHugh KP et al (2014) Mass spectrometry analysis of gingival crevicular fluid in the presence of external root resorption. Am J Orthod Dentofac Orthop 145(6):787–798. https://doi.org/10.1016/j.ajodo.2014.03.013
Carneiro LG, Venuleo C, Oppenheim FG et al (2012) Proteome data set of human gingival crevicular fluid from healthy periodontium sites by multidimensional protein separation and mass spectrometry. J Periodontal Res 47(2):248–262. https://doi.org/10.1111/j.1600-0765.2011.01429.x
Ngo LH, Darby IB, Veith PD et al (2013) Mass spectrometric analysis of gingival crevicular fluid biomarkers can predict periodontal disease progression. J Periodontal Res 48(3):331–341. https://doi.org/10.1111/jre.12012
Uitto V-J (2003) Gingival crevice fluid—an introduction. Periodontol 2000(31):9–11
Embery G, Waddington R (1994) Gingival crevicular fluid: biomarkers of periodontal tissue activity. Adv Dent Res 8(2):329–336
Alfano MC (1974) The origin of gingival fluid. J Theor Biol 47(1):127–136
Carneiro LG, Nouh H, Salih E (2014) Quantitative gingival crevicular fluid proteome in health and periodontal disease using stable isotope chemistries and mass spectrometry. J Clin Periodontol 41(8):733–747. https://doi.org/10.1111/jcpe.12262
Ren Y, Vissink A (2008) Cytokines in crevicular fluid and orthodontic tooth movement. Eur J Oral Sci 116(2):89–97. https://doi.org/10.1111/j.1600-0722.2007.00511.x
Vishwakarma A, Sharpe P, Shi S et al (2014) Stem cell biology and tissue engineering in dental sciences. Elsevier Science, Amsterdam
Wilmes P, Heintz-Buschart A, Bond PL (2015) A decade of metaproteomics: where we stand and what the future holds. Proteomics 15(20):3409–3417. https://doi.org/10.1002/pmic.201500183
Bao K, Belibasakis GN, Selevsek N et al (2015) Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket. Sci Rep 5:15999. https://doi.org/10.1038/srep15999
Barbieri G, Solano P, Alarcon JA et al (2013) Biochemical markers of bone metabolism in gingival crevicular fluid during early orthodontic tooth movement. Angle Orthod 83(1):63–69. https://doi.org/10.2319/022812-168.1
Kereshanan S, Stephenson P, Waddington R (2008) Identification of dentine sialoprotein in gingival crevicular fluid during physiological root resorption and orthodontic tooth movement. Eur J Orthod 30(3):307–314. https://doi.org/10.1093/ejo/cjn024
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422(6928):198–207. https://doi.org/10.1038/nature01511
Andrews GL, Simons BL, Young JB et al (2011) Performance characteristics of a new hybrid triple quadrupole time-of-flight tandem mass spectrometer. Anal Chem 83(13):5442–5446. https://doi.org/10.1021/ac200812d
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372. https://doi.org/10.1038/nbt.1511
Tsuchida S, Satoh M, Umemura H et al (2012) Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics 12(13):2190–2202. https://doi.org/10.1002/pmic.201100655
Bostanci N, Heywood W, Mills K et al (2010) Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MS E (gingival exudatome). J Proteome Res 9(5):2191–2199. https://doi.org/10.1021/pr900941z
Silva-Boghossian CM, Colombo APV, Tanaka M et al (2013) Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS One 8(10):e75898. https://doi.org/10.1371/journal.pone.0075898
Armitage GC (2000) Development of a classification system for periodontal diseases and conditions. Northwest Dent 79(6):31–35
Chen Y-Y, Lin S-Y, Yeh Y-Y et al (2005) A modified protein precipitation procedure for efficient removal of albumin from serum. Electrophoresis 26(11):2117–2127. https://doi.org/10.1002/elps.200410381
Neuhoff V, Arold N, Taube D et al (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9(6):255–262. https://doi.org/10.1002/elps.1150090603
Schmidt C, Lenz C, Grote M et al (2010) Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal Chem 82(7):2784–2796. https://doi.org/10.1021/ac902710k
Shilov IV, Seymour SL, Patel AA et al (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6(9):1638–1655. https://doi.org/10.1074/mcp.T600050-MCP200
Nesvizhskii AI, Keller A, Kolker E et al (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75(17):4646–4658
Liu H, Sadygov RG, Yates JR (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76(14):4193–4201. https://doi.org/10.1021/ac0498563
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. https://doi.org/10.1093/nar/gkn923
Delaville M, Delaville G, Delaville J (1954) Solubility characteristics of the albumin fraction of blood serum in ethyl alcohol-trichloroacetic acid solutions; application to the determination of the various protein fractions of serum (Caractere de solubilite de la fraction albuminique du serum sanguin dans les solutions d'ethanol trichloracetique; application au dosage des diverses fractions proteiques du serum). Ann Biol Clin (Paris) 12(5–6):320–323
Levine S (1954) Solubilization of bovine albumin in nonaqueous media. Arch Biochem Biophys 50(2):515–517
Choi Y-J, Heo S-H, Lee J-M et al (2011) Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci 9(1):42. https://doi.org/10.1186/1477-5956-9-42
Eley BM, Cox SW (1992) Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidyl peptidase IV-like activities in gingival crevicular fluid: a comparison of levels before and after periodontal surgery in chronic periodontitis patients. J Periodontol 63(5):412–417. https://doi.org/10.1902/jop.1992.63.5.412
Travan S, Li F, D'Silva NJ et al (2013) Differential expression of mitogen activating protein kinases in periodontitis. J Clin Periodontol 40(8):757–764. https://doi.org/10.1111/jcpe.12123
Capelli J, Kantarci A, Haffajee A et al (2011) Matrix metalloproteinases and chemokines in the gingival crevicular fluid during orthodontic tooth movement. Eur J Orthod 33(6):705–711. https://doi.org/10.1093/ejo/cjq148
Funding
There was no grant support or funding for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was by the responsible ethics committee (University of Goettingen) with protocol number 23/7/15.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Electronic supplementary material
Supplementary Table 1
(PDF 1323 kb)
Supplementary Table 2
(PDF 1246 kb)
Rights and permissions
About this article
Cite this article
Batschkus, S., Cingoez, G., Urlaub, H. et al. A new albumin-depletion strategy improves proteomic research of gingival crevicular fluid from periodontitis patients. Clin Oral Invest 22, 1375–1384 (2018). https://doi.org/10.1007/s00784-017-2213-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-017-2213-0