Abstract
Objectives
Although the importance of the epigenetic changes in tumors, including oral squamous cell carcinomas (OSCCs), is now becoming apparent, the mechanisms that trigger or cause aberrant DNA methylation in cancer are still unrevealed. DNA methylation is regulated by a family of enzymes, DNA methyltransferases (DNMTs). DNMT gene expression analysis, as well as genetic polymorphisms, has not been previously evaluated in OSCC.
Materials and methods
In 65 OSCC patients, SYBR Green real-time PCR method was assessed for relative quantification of DNMT1, DNMT3A, and DNMT3B mRNAs, normalized to TATA-binding protein (TBP) mRNA. The expression levels of all three genes were dichotomized as high or low, with a twofold change of normalized mRNA expression used as the cutoff value. Polymorphisms in DNMT1 (rs2228612) and DNMT3B (rs406193) were analyzed in 99 OSCCs by TaqMan SNPs genotyping assays.
Results
DNMT1, DNMT3A, and DNMT3B were overexpressed in 36.9, 26, and 23 % of the OSCC patients, respectively. DNMT1 overexpression was significantly associated with the overall survival, p = 0.029, and relapse-free survival of OSCC patients, p = 0.003. Patients with DNMT1 overexpression, as an independent prognostic factor, had a 2.385 times higher risk to relapse than those with lower expression. The DNMT1 A201G gene polymorphism was associated with a reduced overall survival in OSCC patients, p = 0.036.
Conclusions
Our results suggest that DNMT1 could play an important role in modulating OSCC patient survival.
Clinical relevance
DNMT gene expression could be a potential prognostic marker that might lead to an improvement in diagnosis, prognosis, and prospective use of epigenetic-targeted therapy of OSCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral squamous cell carcinoma (OSCC), a subgroup of head and neck squamous cell carcinoma (HNSCC), is a common malignant tumor characterized by a high incidence of nodal metastasis, high recurrence rate, and poor survival [1]. Oral carcinogenesis develops through a multistage process that results from the accumulation of both genetic and epigenetic alterations of tumor-associated genes [1–3]. Epigenetic modifications are heritable changes that modulate gene expression without a change in the DNA sequence itself. The key epigenetic modification in mammalian genome is the DNA methylation of cytosine located 5′ to guanosine in a CpG dinucleotide. Aberrant methylation of CpG islands, CG-rich regions within the promoter regions, causes transcriptional silencing of tumor suppressor genes and malignant transformation [4]. Aberrantly methylated tumor suppressor genes have been suggested as a potential diagnostic, prognostic, and/or predictive tool in a variety of tumors, including OSCC [3, 5–8]. However, due to tissue-type and cell-type specific signature of aberrant DNA methylation in tumors, it is difficult to identify a specific methylation panel as diagnostic and/or prognostic biomarkers for each cancer type.
The covalent addition of a methyl group to cytosine residue of CpGs is catalyzed by a family of enzymes DNA methyltransferases (DNMTs), in the presence of cofactor SAM (S-adenosyl methionine), as a methyl group donor [4]. DNMT1 is mainly involved in the maintenance of DNA methylation after replication and catalyzes the transfer of the methyl group to the daughter DNA strand due to its 10–50-fold higher binding affinity to hemimethylated DNA [9]. DNA methyltransferases DNMT3A and DNMT3B interact with transcription machinery and mediate de novo methylation [4]. Evidences show that DNMT1 may also exert de novo CpG island methylation [4, 10] and/or interact with DNMT3A and DNMT3B in de novo methyltransferase activity, as well as with histone modifying enzymes, and methyl-CpG binding proteins [11].
Overexpression of DNMTs has been reported in a variety of tumors, including breast, gastric, hepatocellular, lung, pancreatic, and head and neck carcinomas [12–22]. It has been suggested that DNMTs may be involved in establishing aberrant DNA methylation patterns in cancer. Due to its ubiquitous overexpression in a number of epithelial cancers, DNMT mRNA and/or protein overexpression could potentially be used as a general tumor molecular marker [13]. In that way, the necessity to create a specific methylation panel as a diagnostic and/or prognostic tool for each cancer type could be significantly reduced.
The current study is the first one investigating the mRNA expression of DNMT1, DNMT3A, and DNMT3B in OSCC and their association with the clinical outcome. While only a few studies examined DNMT polymorphisms and their prognostic or risk association in HNSCCs [23–25], in oral cancer, these polymorphisms have not been investigated. The pre-specified hypotheses tested in the current study was that DNMT expression level and polymorphisms could potentially be markers of overall survival (OS) and/or relapse-free survival (RFS) in OSCC. Thus, analysis of DNMT gene expression could lead to an improvement in diagnosis and prognosis to identify OSCC patients who would benefit from more aggressive therapy. In addition, specific epigenetic and gene expression signatures could be of great value for predicting response to epigenetic therapy of HNSCC and OSCC.
Materials and methods
Subjects
This study was performed in accordance with the Declaration of Helsinki, following the Ethics Committee approval given by the Military Medical Academy, Belgrade. All the participants in the study were randomly selected Caucasians with the same ethnicity, diagnosed with OSCC that underwent an operation at the Clinic for Maxillofacial Surgery, Military Medical Academy, Belgrade, Serbia, between 2005 and 2010. The patients were not given adjuvant chemotherapy or radiation prior to surgery, and subsequently all patients were treated with radiotherapy. Histologically confirmed OSCCs of the tongue and the floor of the mouth were staged according to the TNM classification system for oral and oropharyngeal tumors [26]. The basic demographic characteristics (age, sex, smoking, and alcohol consumption), as well as clinical characteristics (histological and nuclear grade, stage, tumor size, nodal status, and recurrences appearance), are summarized in Table 1. The median patient age was 58 years, range 36–80 years, Table 1. Polymorphisms in DNMT1 (A201G, rs2228612, Ile311Val) and DNMT3B (C501T, rs406193, intergenic) were analyzed from the genomic DNA isolated from 99 tumor tissue of OSCC patients, of which 20 were WHO stage II tumors and 79 were stage III tumors, Table 1. Based on the quality of RNA, 65 cases were selected for the expression analysis of DNMT1, DNMT3A, and DNMT3B genes. Of the 65 tumors, 13 were WHO stage II tumors and 52 were stage III tumors. The recurrence rate was high, 68 % (44/65), and from a total of 38 patient deaths, 37 were preceded by a tumor recurrence (p = 0.000). Reported recommendations for tumor marker prognostic studies (REMARK) criteria were followed throughout this study [27]. All analyses were performed blinded to the study endpoint.
DNA isolation and DNMT1 and DNMT3B genotyping
DNA was extracted from 99 snap-frozen oral cancer tissue samples using the TRIZOL Reagent (Invitrogen, France). The screening for single nucleotide polymorphisms (SNPs) within the DNMT1 (A201G, rs2228612) and DNMT3B gene (C501T, rs406193) was carried out by TaqMan SNPs genotyping assays (Applied Biosystems, UK), according to the manufacturer’s protocols. The SNP genotyping was performed blinded to patient status and expression analysis.
RNA isolation and real-time PCR
RNA was extracted from 65 fresh-frozen tissue samples, preserved at −80 °C, using TRIZOL Reagent (Invitrogen, France), according to the manufacturer’s instructions. The cDNA was prepared using Superscript II RNase H-reverse transcriptase (Invitrogen, France), following the manufacturer’s instructions. Specific DNMT1, DNMT3A, and DNMT3B mRNA expression was assessed by semi-quantitative real-time PCR with SYBR-Green master mix on a 7500 real-time PCR system (Applied Biosystems, UK). The real-time PCR reaction for each sample was duplicated, and the average value was used in the further analysis. The specific primer sets for DNMTs were previously described by Girault et al., where the amplification of contaminating genomic DNA was avoided by placing the one of the two primers at the junction between two exons, or in the different exons [12]. The samples were normalized to TATA-binding protein (TBP), a component of the DNA-binding protein complex transcription factor IID. The TBP was used as an endogenous control for normalization, instead of 18S rRNA or β-actin, to correct for potential variations in the amounts of RNA as DNMT1 gene expression is proliferation-dependent and has a role in maintaining DNA methylation patterns during replication [12]. The relative target gene expression level was also normalized to a calibrator, consisting of a pool of normal oral tissue specimens. Results, expressed as N-fold differences in target gene expression relative to the TBP gene and the calibrator, were determined by the formula N target = 2(ΔCt calibrator − ΔCt sample), where ΔCt values of the calibrator and the sample were calculated by subtracting the average Ct value of the target gene from the Ct value of the TBP gene [12]. Relative expression ratios were calculated using the Pfaff mathematical model, based on the PCR efficiencies, as described previously [28].
Statistical analysis
Statistical analysis was assessed by SPSS 20.0 software (IBM, USA). Clinical, patho-histological, and etiological parameters were compared using the χ2 test or Fisher’s exact test. Spearman’s rank correlation test was used to evaluate relationships between continuous variables.
To validate the efficacy of DNMT overexpression to discriminate outcomes, we summarized the data in a receiver operating characteristic (ROC) curve. The ROC curve plotted the sensibility (true positives) against 1—specificity (false positives), considering each value as a possible cutoff value. The area under the curve (AUC) gave an overall measure to discriminate efficacy of a molecular marker.
The overall survival (OS) and relapse-free survival (RFS) were estimated by using the Kaplan-Meier survival curves, and the significance of differences between survival rates was determined using the log-rank test. The OS was calculated from the time of surgery until death from any cause or last follow-up. RFS were calculated from the time of surgery until the first observation of any recurrence or death due to any cause. If a patient had no recurrences or died, RFS was censored at the time of the last follow-up. Hazard ratios (HR) for OS and RFS were estimated by Cox proportional hazard regression analysis, with 95 % confidence interval (95 % CI). Variables that were significant or near-significant (p < 0.200) in univariate analysis were selected to be included in the multivariate analysis, according to previous studies [8, 22]. Multivariate analysis was used to identify independent prognostic factors of OS and RFS using the Cox proportional hazards model. The Cox model was performed using the forward stepwise method that removed variables with p < 0.100. All the analyses were two-sided, and associations were considered significant with p values less than 0.05 (p < 0.05).
Results
DNMT1, DNMT3A, and DNMT3B mRNA expression analysis
Relative expression levels of DNMT1, DNMT3A, and DNMT3B mRNAs were assessed by semi-quantitative real-time PCR with SYBR Green, normalized to the Ct value of the reference gene TATA-binding protein, TBP. The prognostic performance of these three studied genes was assessed using ROC-AUC analysis. In short, if a molecular marker has a very strong discriminative value, the ROC curve will be close to the upper left corner and the AUC will be close to 1.0; generally good molecular markers have a value between 0.7 and 0.9, as opposed to the markers with poor discriminative power, where the ROC curve will lie close to the diagonal and the AUC will be close to 0.5. DNMT1 emerged as the most discriminatory marker of poor outcome (death) in the OSCC (ROC-AUC, 0.763, p = 0.000), compared to other DNMTs, Table 2, Supplement Fig. 1A. DNMT3A and DNMT3B mRNA levels were similar between the subgroups of patients with poor and good outcomes, with ROC-AUC values close to 0.5 (ROC-AUC, 0.545 and 0.572, respectively), suggesting that overexpression of these genes is not a major prognostic marker of poor outcome in OSCC.
In the absence of a clinically defined cutoff point for DNMT overexpression, to minimize confounding bias, we have examined several options, such as mean, median, arbitrary twofold change, and cutoff values assessed by ROC analysis curve. Initially, the ROC-AUC analysis was used to validate the optimal cutoff point for DNMT mRNA overexpression. An ROC curve demonstrated that for DNMT1, DNMT3A, and DNMT3B expression, fold change values of 1.742, 1.46, and 1.58, respectively, could be potential cutoff points. According to the cutoff value for DNMT1 expression of 1.742, Supplement Fig. 1A, the Cox hazard regression analysis revealed that patients with DNMT1 expression higher than 1.742 have a 3.09 times higher risk of poor outcome compared to patients with lower expression of mRNA, HR = 3.09 (1.56–6.15, 95 % CI, p = 0.000), Supplement Fig. 1B. However, as previously indicated, a fold difference in DNMT1 expression less than at least twofold may result from the imprecise nature of semi-quantitative RT-PCR [19]. Therefore, after examining several options and ROC and Kaplan-Meier analysis to determine the prognostic potential of DNMT1 in oral cancer, the cutpoint for DNMT genes’ overexpression for predicting patient death was defined as greater than or equal to twofold gene expression change of normalized mRNA. The defined overexpression was in accordance with the mean values of DNMT1, DNMT3A, and DNMT3B normalized mRNA levels (1.993, 1.760, and 1.921, respectively), Table 2. Considering this twofold change as a cutoff point, we reanalyzed the relation between patient survival and elevated DNMT1 expression and obtained a test sensitivity of 50 %, specificity of 81.48 %, positive predictive value of 79.17 %, negative predictive value of 53.66 %, and accuracy of 82.6 %. Of the 65 OSCC patients, 24 had DNMT1 mRNA levels above the defined cutoff (36.9 %), DNMT3A mRNA was overexpressed in 17 (26 %) and DNMT3B in 15 (23 %) of OSCC patients.
Association of DNMT1, DNMT3A, and DNMT3B mRNA expression with the clinicopathological data, the overall survival (OS), and the relapse-free survival (RFS)
An association of DNMT mRNA overexpression with clinicopathological features is presented in Table 1. The elevated mRNA levels of DNMTs were not significantly associated with gender, age, smoking status, or alcohol use. Higher DNMT3B mRNA levels were found to be more prevalent in females than in males, p = 0.092. The elevated mRNA levels of DNMT1 were more frequent in advanced histological and nuclear grade OSCC cases, p = 0.012 and p = 0.039, respectively, Table 1. The overall prevalence of overexpressed DNMT3B mRNA was more frequent in advanced stage III compared to stage II, but this difference did not reach statistical significance, p = 0.065. In the studied OSCC cohort, as shown in Table 1, the DNMT1 overexpression showed a tendency toward association with relapses, p = 0.055. No significant correlation was found between the co-expression of DNMTs and clinicopathological data, neither did the expression of DNA methyltransferase genes correlate with each other.
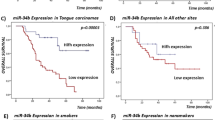
The mean and median follow-up time in studied OSCC cohort were 33.7 months and 30 months, respectively. There have been no clinical data of deaths from other causes. According to the results of the Kaplan-Meier survival analysis, OSCC patients with high DNMT1 mRNA expression had significantly worse overall survival, p = 0.029, Fig. 1a. The Cox hazard regression analysis revealed that patients with DNMT1 overexpression have a 1.99 times higher risk of poor outcome compared to patients with lower expression of mRNA, HR = 1.990 (1.051–3.770, 95 % CI, p = 0.035), Table 3. Variables with univariate significance (p < 0.200) were included in the multivariate analysis using the Cox proportional hazards model for OS and RFS, Table 3. However, in multivariate Cox’s regression analysis, contribution of DNMT1 expression did not persist as an independent prognostic factor, Table 4. Kaplan-Meier survival analysis revealed that DNMT1 overexpression was significantly associated with RFS of OSCC patients (log-rank test, p = 0.003), Fig. 1b. Moreover, the Cox proportional hazards regression analysis showed that the DNMT1 mRNA overexpression was an independent prognostic factor for poor relapse-free survival, HR = 2.385 (1.310–4.341), p = 0.004, Table 4.
Survival distributions, estimated by the Kaplan-Meier method, for DNMT1 mRNA expression analysis (a, b) and DNMT1 A201G, rs2228612 polymorphism (c). a Overall survival (OS) and b relapse-free survival (RFS) curves stratified by DNMT1 mRNA expression in 65 OSCC patients. DNMT mRNA overexpression is defined as greater than or equal to twofold gene expression of normalized mRNA. Patients with OSCCs overexpressing the DNMT1 gene (high expression) had significantly lower 5-year OS (p = 0.029, a) and RFS (p = 0.003, b) than those without DNMT1 overexpression (low expression). c Overall survival curves for DNMT1 A201G, rs2228612 polymorphism in 99 OSCC patients. Patients with the AG genotype (13 out of 99 OSCCs) had the reduced overall survival (p = 0.036, c)
Association of DNMT1 (A201G, rs2228612) and DNMT3B (C501T, rs406193) gene polymorphisms with the clinicopathological features and survival
Polymorphisms in DNMT1 (A201G, rs2228612) and DNMT3B (C501T, rs406193) were analyzed in 99 OSCCs. Our analyses did not show a statistically significant correlation between the DNMT1 and DNMT3B SNPs and corresponding clinical parameters, Table 1. Survival analysis revealed that OSCC patients with AG genotype of DNMT1 A201G gene polymorphism (13 out of 99 OSCCs) had the reduced overall survival, p = 0.036, Fig. 1c. In a subset of 65 OSCC cases, the simultaneous analysis of genotype and expression data did not show a statistically significant correlation between studied DNMT1 and DNMT3B polymorphisms and corresponding mRNA expression (data not shown).
Discussion
Over the last decade, there has been an increase of potential clinical implications of DNA methylation-based biomarkers as potential diagnostic, prognostic, and/or predictive tools in a variety of tumors, including OSCC [2, 4, 5]. Although the importance of the epigenetic changes in a variety of human tumors is now becoming apparent, the mechanisms that trigger or cause aberrant DNA methylation in cancer are still unrevealed. The DNA methyltransferases (DNMTs) are the enzymes responsible for the covalent addition of the methyl group on CpG sites. Although DNMTs were originally classified as maintenance (DNMT1) or de novo DNMTs (DNMT3A and DNMT3B), probably all three DNMTs cooperate and possess both de novo and maintenance roles in establishing DNA methylation [4]. Since DNA methylation panels can provide a better information than a single marker, previous studies evaluated a large number of DNA methylation markers [29, 30]. Although this multi-marker approach is now widely used, the multiple methylation marker screening does not represent a cost-effective and practical approach. It is difficult to identify a specific methylation panel, to use it in large-scale cancer studies or a routine clinical setting, and also challenging to interpret. Moreover, each tumor type display differences in DNA methylation patterns, and differences may be tissue-type, cell type-specific, or even inter-individual [31]. Thus, it has been suggested that DNMT overexpression could potentially be used as a general tumor molecular marker, due to its involvement in establishing aberrant DNA methylation during cancerogenesis [13] and to its ubiquitous overexpression in various epithelial cancers [12–15]. In that way, the necessity to create a specific methylation panel as a diagnostic and/or prognostic tool for each cancer type could be significantly reduced. However, DNMT mRNA expression and genetic polymorphisms have not been previously determined in the OSCC.
Our results indicate, for the first time, that DNMT1 mRNA overexpression could be a potential predictor of poor clinical outcome (p = 0.029) and prognostic molecular marker of reduced relapse-free survival (p = 0.003) in OSCC patients. The current study showed that DNMT1 overexpression is an independent marker of reduced RFS, and patients with DNMT1 overexpression had a 2.385 times higher risk to relapse than those with lower expression. We have also demonstrated that genetic variation in the DNMT1 gene (A201G, rs2228612) is associated with reduced overall survival in oral cancer patients (p = 0.036).
Our results are consistent with previous findings of elevated mRNA levels of DNMT1 shown to be an independent prognostic factor in non-small cell lung carcinomas (NSCLC) [14, 19]. Furthermore, DNMT1 mRNA was overexpressed and correlated with lymph node metastasis in esophageal carcinomas [32].
A central role in evaluating diagnostic or prognostic ability of biomarker to discriminate the true state or outcome of patients is finding of the optimal cutoff values. The use of arbitrary cutoff points is the least accurate method, since cutoffs may be either too low (eliminating positive results) or too high (increasing false-positive results). ROC curve analysis has been used extensively for the assessment of diagnostic or prognostic ability of markers [33]. In the absence of a clinically defined cutoff point for DNMT overexpression, we have examined several options to dichotomize the expression levels of the studied three genes as high (overexpressed) or low. According to the cutoff value assessed by ROC analysis curve, the hazard regression analysis revealed that patients with DNMT1 mRNA expression higher than 1.742-fold change have a 3.09 times higher risk of poor outcome compared to patients with lower expression of mRNA (p = 0.000), Supplement Fig. 1A and B. However, even though the DNMT1 shows the constitutive low level of mRNA expression in normal cells, and only modest overexpression in tumors and with lower frequency [9, 34], it was previously suggested that a fold difference in DNMT1 expression less than twofold may result from the imprecise nature of semi-quantitative RT-PCR [19]. Furthermore, a previous study, which compared the mRNA levels between semi-quantitative RT-PCR and quantitative real-time PCR, indicated that the expression levels of DNMTs measured by semi-quantitative RT-PCR may not be sufficient for studying an association between DNMT expression and patient survival at a different cutoff value (1.5-fold, twofold, threefold) [19]. Therefore, after examining all options to determine the prognostic potential of DNMT1 mRNA expression in oral cancer, the cutoff value for DNMT overexpression for predicting patient poor outcome was defined as greater than or equal to twofold gene expression change of normalized mRNA. In addition, the defined twofold change was in accordance with the mean value of DNMT1 mRNA expression fold change (1.993). Considering this twofold change as a cutoff point, we reanalyzed the relation between patient survival and elevated DNMT1 expression. Nevertheless, we also observed a significant association between elevated DNMT1 mRNA levels with poor clinical outcome (p = 0.029) and reduced relapse-free survival (p = 0.003) in OSCC patients. Further studies in a large cohort are needed to understand the biologic significance of a fold change at a different cutoff value and the effect of a different threshold of DNMT1 overexpression on patient survival.
Previously, DNMT1 mRNA and protein overexpressions were associated with poor prognosis in laryngeal carcinomas [20]. Positive immuno-histochemical staining for DNMT1 in pharyngeal cancer cells and clinical samples was significantly associated with a poor response to therapy and shorter patient survival [21], indicating that DNMT1 may be a significant clinical predictor in HNSCCs. Furthermore, increased DNMT1 protein expression correlated with tumor size, histologic differentiation, and tumor stage of OSCC [35]. However, in another study, DNMT1 and DNMT3B protein expression was not higher in OSCC than in oral leukoplakias or controls, while DNMT3A was overexpressed in OSCC compared to controls, but not for oral leukoplakias [36]. A potent DNA methyltransferase inhibitor, zebularine, significantly reduced viability and DNA synthesis of treated head and neck cancer cells, by induction of cell cycle arrest and apoptosis [37], and suppressed the 5-fluorouracil-induced apoptosis in oral cancer cells [38]. These findings indicate that DNMT1 overexpression could have a substantial effect on cancer progression and clinical outcome and might be a potential treatment target in HNSCC and OSCC.
The molecular mechanism of DNMT1 survival modulation remains to be elucidated. It has been suggested that DNMT1 could play an essential role in cancer cell proliferation and cell survival [39], in addition to its role in the maintenance of DNA methylation [9, 14, 18]. DNMT1 interacts with proteins found at DNA replication forks (PCNA), proteins associated with cell cycle regulation or response to DNA damage (p21, Rb protein, p53), and with a number of proteins directly involved in the epigenetic control of gene expression, including DNMT3A and DNMT3B, and histone deacethylase (HDAC1, HDAC2) [9]. Antisense oligonucleotides targeting DNMT1 and DNMT3B had pronounced anti-proliferative effects in esophageal, lung, and malignant pleural mesothelioma cancer cells [40]. In addition, transfection of DNMT1 small interfering RNA (siRNA) decreased DNMT1 protein levels and led to the increase of p21 and suppression of cell proliferation in head and neck cancer cell line [41]. DNMT1 silencing vector reduced tumor growth, reduced invasion, and attenuated treatment resistance in bladder cancer cells [42]. The treatment of colon cancer cells with interleukin (IL)-6, a major effector cytokine of inflammation, resulted in an increase in DNMT1 expression and DNA methylation of tumor-associated genes [43], which indicated that DNMT1 could also have an important role in inflammation-associated carcinogenesis.
Our study showed that the variant homozygote genotype of the DNMT1 polymorphism A201G (rs2228612) reduces the overall survival in our oral cancer cohort (p = 0.036). The GG homozygote variant genotype of the studied DNMT1 A201G polymorphism was previously associated with the increased breast cancer susceptibility in Han Chinese women [44], while the opposite finding of breast cancer risk reduction was reported in female Caucasian patients [45]. The studied DNMT1 A201G polymorphism (I311V) is located in the N-terminal part of the gene, encoding the nuclear localization signal (NLS) (1–343 amino acid residues), and a PCNA-interacting region [9, 46]. The N-terminus of a protein often has a substantial influence on protein stability and accumulation. Although deletion of the N-terminal 120 amino acids significantly increased the DNMT1 protein stability and subsequent accumulation in breast cancer cells, deletion of the NLS domain containing the studied DNMT1 A201G polymorphism did not exert a further effect on DNMT1 protein accumulation [15]. Nevertheless, the variants in the N-terminal part of DNMT1 could be essential for the efficient catalytic activity of the enzyme and the preference for hemimethylated sites, as well as for its interactions with other proteins [9, 46]. Previously, studied DNMT1 A201G polymorphism was related to lower levels of global DNA methylation in autoimmune thyroid disease [47]. Thus, this non-synonymous I311V substitution could transform the structure of the NLS domain and decrease the DNA-binding ability of the enzyme, consequently reducing the global DNA methylation levels.
In conclusion, the current study is the first one demonstrating that the DNMT1 mRNA overexpression and genetic variation could be associated with survival in oral cancer patients. Thus, OSCC patients who would benefit from aggressive therapy could be identified. Although the sample size in the current study is sufficiently large for oral cancer, further studies are warranted to be conducted in expanded patient cohorts to verify these associations for different cancer types. In addition, further studies are needed to assess associations between DNMT overexpression and DNMT polymorphisms with DNA methylation. Furthermore, there is a growing interest in DNA methylation-based therapy due to the reversibility of epigenetic changes, and various DNMT inhibitors exhibit promising results in anti-tumor therapy, but have yet to be explored in OSCC. Different strategies for oral cancer therapy via knockdown or DNMT inhibition are to be evaluated. Thus, the analysis of DNMT gene expression could potentially lead to an improvement in diagnosis, prognosis, and prospective use of epigenetic-targeted therapy of HNSCC and OSCC.
References
Scully C, Bagan J (2009) Oral squamous cell carcinoma overview. Oral Oncol 45(4–5):301–308
Mascolo M, Siano M, Ilardi G, Russo D, Merolla F, De Rosa G, Staibano S (2012) Epigenetic disregulation in oral cancer. Int J Mol Sci 13(2):2331–2353
Shaw R (2006) The epigenetics of oral cancer. Int J Oral Maxillofac Surg 35(2):101–108
Esteller M (2007) Epigenetic gene silencing in cancer: the DNA hypermethylome. Human Molecular Genetics. 16:R50–R59
Deng D, Liu Z, Du Y (2010) Epigenetic alterations as cancer diagnostic, prognostic, and predictive biomarkers. Adv Genet 71:125–176
Delpu Y, Cordelier P, Cho W, Torrisani J (2013) DNA Methylation and cancer diagnosis. Int J Mol Sci 14:15029–15058
Supic G, Kozomara R, Brankovic-Magic M, Jovic N, Magic Z (2009) Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol 45:1051–1057
Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z (2011) Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol 47:702–708
Kar S, Deb M, Sengupta D, Shilpi A, Parbin S, Torrisani J, Pradhan S, Patra S (2012) An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 7(9):994–1007
Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, Baylin SB, Schuebel KE (2006) De novo CpG island methylation in human cancer cells. Cancer Res 66(2):682–692
RK L, YC W (2014) Dysregulated transcriptional and post-translational control of DNA methyltransferases in cancer. Cell Biosci 4:–46
Girault I, Tozlu S, Lidereau R, Bieche I (2003) Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Canc Res 9:4415–4422
Mutze K, Langer R, Schumacher F, Becker K, Ott K, Novotny A, Hapfelmeier A, Höfler H, Keller G (2011) DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer 47(12):1817–1825
Xing J, Stewart D, Gu J, Lu C, Spitz M, Wu X (2008) Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer 98:1716–1722
Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG (2005) Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 280(18):18302–18310
Ding WJ, Fang JY, Chen XY, Peng YS (2008) The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig Dis Sci 53:2083–2089
Saito Y, Kanai Y, Nakagawa T, Sakamoto M, Saito H, Ishii H, Hirohashi S (2003) Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer 105(4):527–532
Gao J, Wang L, Xu J, Zheng J, Man X, Wu H, Jin J, Wang K, Xiao H, Li S, Li Z (2013) Aberrant DNA methyltransferase expression in pancreatic ductal adenocarcinoma development and progression. J Exp Clin Cancer Res 32:86
Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH (2006) Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer 107(5):1042–1049
Wang J, Xu Y, Li J, Sun X, Wang LP, Ji WY (2012) The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation in laryngeal carcinoma. Oral Oncol 48(6):541–546
Chen CC, Chen WC, Wang WH, Lu CH, Lin PY, Lee KD, Chen MF (2011) Role of DNA methyltransferase 1 in pharyngeal cancer related to treatment resistance. Head Neck. 33(8):1132–1143
Bieche I, Tozlu S, Girault I, Lidereau R (2004) Identification of a three-gene expression signature of poor-prognosis breast carcinoma. Mol Cancer 3:37
Zhu J, Du S, Zhang J, Wang Y, Wu Q, Ni J (2015) Polymorphism of DNA methyltransferase 3B -149C/T and cancer risk: a meta-analysis. Med Oncol 32(1):399
Wang L, Rodriguez M, Kim ES, Xu Y, Bekele N, El-Naggar AK, Hong WK, Mao L, Oh Y (2004) A novel C/T polymorphism in the core promoter of human de novo cytosine DNA methyltransferase 3B6 is associated with prognosis in head and neck cancer. Int J Oncol 25(4):993–999
Liu Z, Wang L, Wang LE, Sturgis EM, Wei Q (2008) Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Lett 268(1):158–165
Warnakulasuriya S, Johnson NW, van der Waal I (2007) Nomenclature and classification of potentially malignant disorders of the oral mucosa. J Oral Pathol Med 36(10):575–580
Altman DG, McShane LM, Sauerbrei W, Taube SE (2012) Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med 9:e1001216
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 29(9):e45
Shaw R, Hall G, Lowe D, Bowers NL, Field JK, Risk JM, et al. (2007) CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol 43:878–886
Marsit CJ, Houseman EA, Christensen BC, Eddy K, Bueno R, Sugarbaker DJ, et al. (2006) Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res 66:10621–10629
Li X, Baker-Andresen D, Zhao Q, Marshall V, Bredy TW (2014) Methyl CpG binding domain ultra-sequencing: a novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes Brain Behav. 13(7):721–731
Zhao SL, Zhu ST, Hao X, Li P, Zhang ST (2011) Effects of DNA methyltransferase 1 inhibition on esophageal squamous cell carcinoma. Dis Esophagus 24(8):601–610
Hajian-Tilaki K (2013) Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med 4(2):627–635
Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (1999) The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 27(11):2291–2298
Shiah SG, Chang LC, Tai KY, Lee GH, Wu CW, Shieh YS (2009) The involvement of promoter methylation and DNA methyltransferase-1 in the regulation of EpCAM expression in oral squamous cell carcinoma. Oral Oncol 45(1):e1–e8
Daniel F, Rivero ER, Modolo F, Lopes TG, Salum FG (2010) Immunohistochemical expression of DNA methyltransferases 1, 3a and 3b in oral leukoplakias and squamous cell carcinomas. Arch Oral Biol 55(12):1024–1030
Napso T, Fares F (2014) Zebularine induces prolonged apoptosis effects via the caspase-3/PARP pathway in head and neck cancer cells. Int J Oncol 44(6):1971–1979
Suzuki M, Shinohara F, Endo M, Sugazaki M, Echigo S, Rikiishi H (2009) Zebularine suppresses the apoptotic potential of 5-fluorouracil via cAMP/PKA/CREB pathway against human oral squamous cell carcinoma cells. Cancer Chemother Pharmacol 64(2):223–232
Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G (2006) Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A 103:14080–14085
Kassis ES, Zhao M, Hong JA, Chen GA, Nguyen DM, Schrump DS (2006) Depletion of DNA methyltransferase 1 and/or DNA methyltransferase 3b mediates growth arrest and apoptosis in lung and esophageal cancer and malignant pleural mesothelioma cells. J Thorac Cardiovasc Surg 131(2):298–306
Oridate N, Lotan R (2005) Suppression of DNA methyltransferase 1 levels in head and neck squamous carcinoma cells using small interfering RNA results in growth inhibition and increase in Cdk inhibitor p21. Int J Oncol 26(3):757–761
Wu CT, Wu CF, Lu CH, Lin CC, Chen WC, Lin PY, Chen MF (2011) Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer 117(22):5221–5233
Foran E, Garrity-Park M, Mureau C, Newell J, Smyrk T, Limburg P, Egan L (2010) Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res 8(4):471–481
Sun MY, Yang XX, Xu WW, Yao GY, Pan HZ, M L (2012) Association of DNMT1 and DNMT3B polymorphisms with breast cancer risk in Han Chinese women from South China. Genet Mol Res 11(4):4330–4341
Kullmann K, Deryal M, MF O, Schmidt W, Mahlknecht U (2013) DNMT1 genetic polymorphisms affect breast cancer risk in the central European Caucasian population. Clin Epigenetics 5(1):7
Fatemi M, Hermann A, Gowher H, Jeltsch A (2002) Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem 269:4981–4984
Arakawa Y, Watanabe M, Inoue N, Sarumaru M, Hidaka Y, Iwatani Y (2012) Association of polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR genes with global DNA methylation levels and prognosis of autoimmune thyroid disease. Clin Exp Immunol 170(2):194–201
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This study was performed in accordance with the Declaration of Helsinki, following the Ethics Committee approval given by the Military Medical Academy, Belgrade, Serbia.
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
The work was supported by the Military Medical Academy, Belgrade, Serbia, grant No. MFVMA/14/12-14.
Informed consent
Written informed consent was obtained from all participants of this study.
Electronic supplementary materials
Supplement Fig. 1.
ROC and Kaplan-Meier analysis performed to determine the DNMT1 prognostic potential in oral cancer. (A) ROC analysis curve of DNMT1 mRNA expression as positive marker for the overall survival outcome; ROC, Receiver-Operating Characteristic, according to poor outcome (death) in the total number of 65 OSCC cases. (B) Kaplan-Meier analysis of overall survival for DNMT1 mRNA overexpression, based on a value of 1.742-fold change, as a cutoff point for DNMT1 overexpression. HR indicates a hazard ratio, based on the Cox Proportional Hazards Regression analysis of DNMT1 mRNA expression in 65 patients with OSCCs. (GIF 101 kb)
Rights and permissions
About this article
Cite this article
Supic, G., Kozomara, R., Zeljic, K. et al. Prognostic value of the DNMTs mRNA expression and genetic polymorphisms on the clinical outcome in oral cancer patients. Clin Oral Invest 21, 173–182 (2017). https://doi.org/10.1007/s00784-016-1772-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-016-1772-9