Abstract
Objective
The aim of the study is to compare detection frequency of periodontal pathogens in patients with aggressive/severe chronic periodontitis using pooled plaque samples from the deepest pockets per quadrant/per sextant.
Methods
In 100 patients with aggressive/chronic periodontitis, subgingival plaque was sampled from the deepest pockets per quadrant (MT4) and per sextant (MT6). Plaque samples were taken using two sterile paper points simultaneously. One paper point from each pocket was pooled with the three other paper points of the pockets (MT4). Subsequently, the remaining four paper points were pooled with two paper points from the deepest pockets from the two remaining sextants (MT6). The content of each vial was analyzed with nucleic-acid based methods for Aggregatibacter actinomycetemcomitans, Tannerella forsythia, Porphyromonas gingivalis, Treponema denticola, Prevotella intermedia, Parvimonas micra, Fusobacterium nucleatum, Campylobacter rectus, Eubacterium nodatum, Eikenella corrodens, and Capnocytophaga sp.
Results
The detection frequency of A. actinomycetemcomitans (MT4/MT6) at 22/24 %, T. forsythia at 93/96 %, P. gingivalis at 78/79 %, T. denticola at 88/90 %, P. intermedia at 40/46 %, P. micra at 75/79 %, F. nucleatum at both 99 %, C. rectus at 84/89 %, E. nodatum at 62/65 %, E. corrodens at 80/87 %, and Capnocytophaga sp. at 49/58 % was higher with MT6 than with MT4. None of these differences were statistically significant.
Conclusion
The detection frequency of the investigated periopathogens was statistically insignificant higher with the sampling method MT6 compared with MT4.
Clinical relevance
In daily dental practice, the plaque sampling of the deepest pockets per quadrant seems to be sufficient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
From the approximately 400 bacterial species colonizing periodontal pockets and a further 300 in the rest of the oral cavity [1, 2], some are frequently associated with periodontal destruction. Aggregatibacter actinomycetemcomitans (formerly known as Actinobacillus actinomycetemcomitans) [3], Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola are considered periodontal pathogens. Prevotella intermedia, Parvimonas micra, Fusobacterium nucleatum, Campylobacter rectus, Prevotella nigrescens, Eikenella corrodens, and Capnocytophaga sp. are associated with periodontal disease [4, 5]. Further, A. actinomycetemcomitans has been closely associated with the aetiology of severe periodontal disease: aggressive Periodontitis (AgP) [6–10] and periodontitis as manifestation of Papillon Lefèvre syndrome [11]. A. actinomycetemcomitans is a microaerophilic, facultative anaerobic and Gram-negative coccoid rod belonging to the family of Pasteurellaceae [3]. Periodontal disease associated with A. actinomycetemcomitans in many cases cannot be treated reliably and predictively by mechanical removal of the subgingival biofilm alone [6, 12–17].
Thus, the detection of A. actinomycetemcomitans is a significant factor contributing to the decision whether mechanical anti-infective therapy should be used in conjunction with systemic antibiotics [18, 19]. Further, depending on the microbial complexes that are detected from subgingival plaque, it has been proposed to apply varying antibiotic regimes [20].
According to the joint statement of the German Society for Periodontology (DG PARO) and the German Society for Dental, Oral, and Maxillofacial Medicine (DGZMK), microbiological testing prior to anti-infective therapy is indicated for the following clinical diagnoses: aggressive periodontitis, generalized severe chronic periodontitis, periodontitis exhibiting progressive attachment loss despite thorough treatment, and severe periodontitis associated with systemic diseases (e.g., HIV infection) [21]. Subgingival plaque samples should be collected from the deepest pockets exhibiting signs of activity, i.e., bleeding or suppuration. A microbiological analysis representative for the subgingival microbiota of the whole oral cavity is relevant for adjunctive systemic antibiotic therapy of certain forms of periodontitis. Therefore, due to economic reasons also, the analysis of pooled plaque sampled from several sites is recommended [21].
Recent research has demonstrated that pooled analysis provides at least the same detection rate as separate analysis of the samples from different sites for A. actinomycetemcomitans, P. gingivalis, T. forsythia, and T. denticola [9, 22] as well as for P. intermedia and F. nucleatum [23].
However, also a high variability of results was shown that depended on the number of sampled sites. The higher the number of sites, the higher the detection rate and the less the variability [9, 22, 24]. Sampling six sites instead of four sites increases the effort. Up to now, the comparison of pooled samples from four to six sites has not been investigated.
Thus, in this study, the results of microbiological semiquantitative PCR analyzes of pooled subgingival plaque samples from the deepest sites per quadrant (MT4) should be compared with the results from the pooled sample from the deepest sites per sextant (MT6). To reveal a difference of 20 % between the detection rates of MT4 and MT6 with a type 1 error α < 0.05 and a test power of 80 %, a minimal sample size of n = 70 is required [22]. Thus, a sample size of n = 100 was chosen.
Material and methods
Patients
One hundred patients under periodontal therapy at the Department of Periodontology, Center for Dentistry and Oral Medicine, Johann Wolfgang Goethe-University Frankfurt/Main and a private practice at Frankfurt/Main, Germany, were recruited between October 2007 and August 2011. To be included in this study, the following inclusion criteria had to be fulfilled:
-
Clinical diagnosis of untreated aggressive or generalized severe chronic periodontitis [18, 19, 21]
-
At least 18 years of age
-
At least five teeth per quadrant
-
Informed written consent
The following criteria lead to exclusion from the study:
-
Need for antibiotic prophylaxis in advance of periodontal diagnosis or treatment
-
Antibiotic therapy within the last 6 months or subgingival debridement (nonsurgical or surgical) within the last 12 months before microbiological sampling
For this study, the diagnoses aggressive (AgP) and generalized chronic periodontitis (ChP) were defined as follows:
-
1.
Aggressive periodontitis [25]:
-
Patient is clinically healthy, i.e., systemic diseases predisposing for periodontitis are not reported (e.g., diabetes mellitus)
-
Radiographic bone loss ≥50 % at at least two different teeth
-
Age ≤ 35 years
-
-
2.
Generalized chronic periodontitis:
-
Attachment loss ≥5 mm at more than 30 % of sites
-
Age > 35 years
-
The study had been approved by the Institutional Review Board for Human Studies of the Medical Faculty of the Johann Wolfgang Goethe-University Frankfurt/Main (Study #189/07). All patients gave written informed consent to participate in the study.
Clinical examinations
At six sites per tooth (mesiobuccal, midbuccal, distobuccal, distooral, midoral, mesiooral), probing depths (PD) and vertical clinical attachment levels (PAL-V) were measured using a manual rigid periodontal probe (PCPUNC15, Hu Friedy, Chicago, IL, USA) to the nearest millimeter. The cemento-enamel junction (CEJ) was used as reference for PAL-V measurements. If the CEJ was destroyed by restorative treatment, the margin of the restoration was taken as reference. Bleeding on probing (BOP) was recorded 30 s after probing.
Microbiological examination
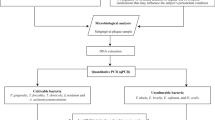
Microbiological sampling was performed within the clinical routine according to the joint statement of the German Society of Periodontology (DG PARO) and the German Society of Dental, Oral, and Maxillofacial Medicine (DGZMK) [21]. For sampling, the four deepest pockets in four different quadrants were selected. The test site was dried by air and held dry using cotton rolls. Simultaneously, two sterile paper points were inserted to the bottom of the respective pocket [9, 22]. After 20 s, the paper points were removed.
One of each paper points was put into one of two transportation vials. Thus, two pooled samples of the same four sites were created. One of the vials was closed (MT4). Then, the deepest pockets of the two remaining sextants were selected—plaque samples were taken and pooled with the other remaining vial (MT6). Hence, for each patient, two transportation vials were loaded: one containing four paper points from the deepest pockets from each quadrant (MT4) and one containing six paper points—four paper points from the deepest pockets from each quadrant and two paper points from the deepest pockets from the two remaining sextants (MT6).
For analysis, a commercially available PCR DNA probe test kit (micro-IDent plus®, Hain Lifescience GmbH, Nehren, Germany) aiming at A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, P. intermedia, P. micra, F. nucleatum, C. rectus, E. nodatum, E. corrodens, and Capnocytophaga sp. was used, which is based on the DNA strip technique. The detection limit of this test is 103 pathogens for A. actinomycetemcomitans and 104 for all other bacteria. The Hain microIDent® plus analysis of the plaque probes was performed in the Laboratories of Hain Lifescience GmbH. First, DNA was extracted from the paper points by adding to every sample 200-μL 5 % Chelex solution, a chelating cation resin suspension used for the rapid extraction of DNA from small biological samples. The samples were then placed in an ultrasonic bath (Branson 5510) at 60 °C for 15 min and finally incubated in a 105 °C thermo block for 15 min. After vortexing and centrifugation at full speed, the supernatant was used for molecular analysis. PCR amplification was carried out according to the micro-IDent® plus manual. From each sample, two separate amplification reactions were performed, the first with the micro-IDent® primer-nucleotide mix for amplification of five species and the second with the micro-IDent® plus primer-nucleotide mix for amplification of the six additional species. The amplification mix (45 μL) consisted of 35 μL of primer–nucleotide mix (micro-IDent® or micro-IDent® plus, respectively), 5 μL of 10× PCR buffer, 2 μL of 25-mM MgCl2, 3 μL of H2O, and 1 U Taq polymerase (MBI Fermentas, Vilnius, Lithuania). Five microliters of DNA solution was added to a final reaction volume of 50 μL. PCR cycling was carried out in a Dual 384-Well GeneAmp® PCR System 9700 (Applied Biosystems Inc., Foster City, CA, USA). The cycling conditions comprised an initial denaturation step at 95 °C for 5 min, 10 cycles at 95 °C for 30 s and at 58 °C for 2 min, 20 cycles at 95 °C for 25 s, at 53 °C for 40 s and at 70 °C for 40 s, and a final extension step at 70 °C for 8 min. Positive and contamination controls were included in each batch of samples. The subsequent reverse hybridization was performed according to the micro-IDent® plus kit. Both amplicons from each sample were separately hybridized to the respective strip, the first coated with the five micro-IDent probes and the second with the six additional micro-IDent® plus probes. Each strip includes two control lines (Conjugate Control, Amplification Control). In short, the biotinylated amplicons were denatured and incubated at 45 °C with hybridization buffer. After PCR products had bound to their respective complementary probe, a highly specific washing step removed any unspecifically bound DNA. Streptavidin-conjugated alkaline phosphatase was added, the samples were washed, and hybridization products were visualized by adding a substrate for alkaline phosphatase. Results could be obtained after approx. 5 h; the “hands-on-time” was about 1.5 h.
Statistical analysis
Two variables were analyzed for each periodontal pathogen:
-
Prevalence, i.e., detection of the pathogen or no detection
-
Semiquantitative classification (groups 0 to 4) (Table 1).
Table 1 Semiquantitative classification of bacterial counts
Detection rate for MT4 and MT6 was compared using the Wilcoxon signed ranks test for paired samples. Agreement of both analyzing strategies was estimated by calculating Cohen’s kappa (detection rate), respectively [26]. Highly positive samples (semiquantitative classification of bacterial counts (Table 1): groups 3 and 4) of MT4 and MT6 were compared with the McNemar chi-squared test.
PD and PAL-V were used to describe the clinical status of sampled sites. All other statistical analyses were done using a PC program (Systat™ for Windows Version 12, Systat Inc. Evanston, IL, USA).
Results
From a total of 100 patients (53 female, 47 male; 45.3 ± 12.3 years of age), clinical and microbiological examinations were obtained. Twenty-nine patients suffered from untreated aggressive and 71 from untreated generalized severe chronic periodontitis (Table 2).
The detection frequency of A. actinomycetemcomitans (MT4/MT6) at 22/24 %, T. forsythia at 93/96 %, P. gingivalis at 78/79 %, T. denticola at 88/90 %, P. intermedia at 40/46 %, P. micra 75/79 %, F. nodatum at both 99 %, C. rectus at 84/89 %, E. nodatum at 62/65 %, E. corrodens at 80/87 %, and Capnocytophaga sp. at 49/58 % was higher with MT6 than with MT4. None of these differences were statistically significant (Table 3). For P. gingivalis, the highly positive samples (semiquantitative classification of bacterial counts: groups 3 and 4) were higher for MT6 than for MT4. For all other tested bacteria, MT4 and MT6 were similar. Analysis failed to detect statistically significant differences between both sampling strategies (Table 3).
Discussion
The detection of A. actinomycetemcomitans has a significant influence on the decision whether or not mechanical anti-infective therapy should be administered in conjunction with systemic antibiotics [27–30]. In this study, a commercially available PCR DNA probe test kit (Hain micro-IDent plus®) was used analyzing the subgingival plaque samples, which is based on the DNA strip technique. Eick et al. compared microbiological cultivation with a commercial PCR-based method for the detection of periodontal pathogens in subgingival plaque. The comparison of the two methods revealed that the micro-IDent kit identified both P. gingivalis and T. forsythia more often than did the cultivation method. Thus, the authors concluded that nucleic acid techniques should replace cultivation methods as gold standard in microbiological diagnosis of progressive periodontitis [31]. Other authors comparing culture and a real-time PCR for detection of periodontal pathogens observed low agreement of both techniques regarding F. nucleatum and P. intermedia [32].
In the present study, paper points were used for microbiological sampling. This is a standard for commercially available bacteriological tests and was also used in many microbiological studies [9, 15, 22, 33]. However, paper points have some disadvantages: If they become wet by gingival crevicular fluid, they lose stiffness making it difficult to move them to the bottom of the pocket. Jervoe-Storm et al. compared the recovery of six periodontal pathogens by paper point samples from different aspects of the lesion (full-length or half-length of the pocket depth) in 20 patients: The authors found out that the recovery of the target pathogens was similar following sampling at various depths of the periodontal lesions [34]. Another disadvantage is that different paper points placed in the same pocket at the same time and for the same period of time may not sample the same microorganisms [22]. Subgingival plaque may be sampled also using curettes [24]. Curettes are made of steel and consequently stay stiff. Further, a curette samples subgingival plaque from a larger area than a paper point. Thus, sampling plaque with a curette might overcome some of the disadvantages of paper points. Jervøe-Storm et al. compared the curette and paper point sampling technique using quantitative real-time PCR [35]. The results demonstrated that the plaque composition with regard to total target pathogens was similar for both sampling techniques. Hence, both techniques seem to be suitable for microbiological diagnostics. Some authors preferred the paper point technique due to reproducibility [36] or reliability [37]. Using paper points of standardized size provides standardized samples which are preferred particularly by the laboratory. Further studies may elucidate to which extent the sampling method affects the variability of microbiological analysis.
Supragingival plaque was not removed before sampling because it had been demonstrated that in periodontally diseased individuals both supragingival and subgingival plaque harbor the targeted periodontal pathogens [24]. Paper points remained 20 s in the periodontal pocket. This procedure has been found to be adequate in studies addressing duration of subgingival plaque sampling [38].
A. actinomycetemcomitans may not be present at all oral sites in a patient suffering from untreated periodontitis. Taking subgingival samples from all teeth would be the most reliable way to detect A. actinomycetemcomitans. However, this method is too time consuming and expensive to be used in daily practice. Sampling of the deepest pocket of each quadrant has been demonstrated to detect quite reliably the subgingival presence of A. actinomycetemcomitans [15] or P. gingivalis [33] in untreated patients. Mombelli et al. sampled and microbiologically analyzed all sites separately and theoretically evaluated different sampling strategies [15, 33]. Yet, their strategies were based on separate analyses of the samples, whereas in daily practice, the different samples taken from the deepest sites per quadrant are pooled prior to analysis for economic reasons. Haffajee et al. analyzed the effect of sampling strategy on the false-negative rate for detection of selected subgingival species. They compared plaque samples obtained from one site per tooth with samples taken from (a) one maxillary first molar, (b) both maxillary first molars, (c) four first molars, (d) six Ramfjord teeth, (e) the deepest pocket, and (f) the four deepest pockets. The highest rates of detected species were found for sampling strategy—(f)the four deepest pockets [38]. Thus, for microbiological analysis in daily practice sampling, the deepest site per quadrant generally is recommended [21].
After sampling, one to six sites per patient and separate analysis per site Beikler et al. reported increasing probability to detect the targeted bacteria with increasing number of sampled sites [24]. However, other authors reported contradicting observations [39]. These differences may be explained by significant differences in methodology: (a) sampling with curettes [24] or paper points [40], and (b) separate [24] or pooled analysis [40]. Thus, at least for pooled analysis up to now, it was not clear whether sampling the deepest sites per sextant (MT6) instead of the deepest per quadrant (MT4) increases the probability of detection. For the PCR, DNA probe test kit that was investigated MT6 failed to demonstrate advantages over MT4.
Another study from our group compared the detection frequency and number of periodontal pathogens in 50 patients showing also untreated aggressive or severe chronic periodontitis [41]: Plaque was also sampled from the deepest pockets per quadrant and per sextant and analyzed for the presence of A. actinomycetemcomitans, P. gingivalis, T. forsythia, and T. denticola. In that study, another analyzing method was used (16S rRNA gene probe), with a detection frequency of 103.3 for A. actinomycetemcomitans and 104 for P. gingivalis, T. forsythia, and T. denticola. The proportion of patients with aggressive periodontitis was 32 %. Detection frequencies and counts for A. actinomycetemcomitans were higher for MT6 than MT4, but the differences reached statistical significance only for the bacterial counts. Detection frequencies and counts were generally high for P. gingivalis, T. forsythia, and T. denticola (>95 %). However, the differences between MT4 and MT6 were not significant.
The studied population in this present study corresponds to the populations analyzed in previous studies [9, 22] regarding patient parameters (age, gender) and clinical parameters (PD, PAL-V). In the present study, A. actinomycetemcomitans was detected in 22 % (MT4) and 24 % (MT6) of all patients, with a proportion of patients with aggressive periodontitis: 29 patients (29 %). There is a body of evidence that the prevalence of A. actinomycetemcomitans is higher in patients with aggressive periodontitis than in patients with chronic periodontitis [9, 22, 42].
For all tested microorganisms, the present study failed to observe statistically significant differences between the semiquantitative bacterial counts of the pooled samples from the deepest pockets from each sextant (MT6) and the semiquantitative bacterial counts from each quadrant (MT4). The detection frequency of A. actinomycetemcomitans, T. forsythia, P. gingivalis, T. denticola, P. intermedia, P. micra, F. nucleatum, C. rectus, E. nodatum, E. corrodens, and Capnocytophaga sp. was higher with MT6 than with MT4. None of these differences were statistically significant. The agreement between both sampling strategies was assessed as Cohen’s kappa [26]. The agreement was moderate (Capnocytophaga sp., E. corrodens and P. micra) to high (A. actinomycetemcomitans, P. gingivalis, T. forsythia, T. denticola, P. intermedia, F. nucleatum, C. rectus, E. nodatum). However, for some patients, MT4 analysis was positive for one of the tested periodontal pathogens, whereas MT6 analysis was not and vice versa. This observation may be explained by an uneven distribution or at least uneven concentrations of the different bacterial species within periodontal pockets—one sampling strategy detects the microorganisms, whereas the other does not. Finally, this might be caused by random chance: One microorganism is located in the eluate which is pipetted in laboratory; the other one is not.
P. gingivalis, T. forsythia, T. denticola, P. micra, F. nucleatum, C. rectus, and E. corrodens were detected in the majority of the patients (detection frequency of approximately 80 % and higher). P. intermedia was prevalent in 46 %, E. nodatum in 65 %, and Capnocytophaga sp. in 58 % of the cases. These detection frequencies are in accordance with observations previously made by others using different microbiological tests in patients with untreated aggressive and generalized severe chronic periodontitis study [9, 22, 43]. Thus, in untreated aggressive and generalized severe chronic periodontitis detection of P. gingivalis, T. forsythia, T. denticola, P. micra, F. nucleatum, C. rectus, and E. corrodens is no essential information because it may be expected in approximately 80 to 100 % of this kind of patients any way.
Within the limitations of the present study, the following conclusion may be drawn: (a) Regarding detection frequency and semiquantitative bacterial counts of the tested microorganisms sampling, the deepest sites per sextant (MT6) have no advantage over sampling the deepest sites per quadrant (MT4). Thus, pooled analysis of subgingival plaque samples from sour sites is as good as from six sites to describe subgingival periodontal pathogens on the patient level (b) P. gingivalis, T. forsythia, T. denticola, P. micra, F. nucleatum, C. rectus, and E. corrodens may be detected in approximately 80 to 100 % of all patients with untreated aggressive and generalized severe chronic periodontitis.
References
Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE (2001) Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783
Paster BJ, Olsen I, Aas JA, Dewhirst FE (2006) The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 42:80–87
Nørskov-Lauritsen N, Kilian M (2006) Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol 56:2135–2146
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr (1998) Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144
van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U (2002) Porphyromonas gingivalis and Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol 29:1023–1028
Bragd L, Dahlen G, Wikström M, Slots J (1985) The capability of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius to indicate progressive periodontitis; retrospective study. J Clin Periodontol 14:95–99
Newman MG, Socransky SS, Savitt ED, Propas DA, Crawford A (1976) Studies of the microbiology of periodontosis. J Periodontol 47:373–379
Tonetti MS, Mombelli A (1999) Early-onset periodontitis. Ann Periodontol 4:39–52
Schacher B, Baron F, Rossberg M, Wohlfeil M, Arndt R, Eickholz P (2007) Aggregatibacter actinomycetemcomitans as indicator for aggressive periodontitis by two analysing strategies. J Clin Periodontol 34:566–573
Haubek D, Ennibi O-K, Væth M, Poulsen S, Kilian M (2008) Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet 371:237–242
Schacher B, Baron F, Ludwig B, Valesky E, Noack B, Eickholz P (2006) Periodontal therapy in siblings with Papillon-Lefèvre syndrome and tinea capitis: a report of two cases. J Clin Periodontol 33:829–836
Christersson L, Fransson C, Dunford R, Zambon J (1992) Microbiological and clinical effects of surgical treatment of localized juvenile periodontitis. J Peridontol 63:465–476
Kornman K, Robertson P (1985) Clinical and microbiological evaluation of therapy of juvenile peridontitis. J Periodontol 56:443–446
Müller H-P, Lange DE, Müller RF (1993) Failure of adjunctive minocycline-HCl to eliminate oral Actinobacillus actinomycetemcomitans. J Clin Periodontol 20:498–504
Mombelli A, Gmür R, Gobbi C, Lang NP (1994) Actinobacillus actinomycetemcomitans in adult periodontitis. I. Topographic distribution before and after treatment. J Periodontol 65:820–826
Takamatsu N, Yano K, He T, Umeda M, Ishikawa I (1999) Effect of initial periodontal therapy on the frequency of detecting Bacteroides forsythus, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J Periodontol 70:574–580
Ehmke B, Moter A, Beikler T, Milian E, Flemmig TF (2005) Adjunctive antimicrobial therapy of periodontitis: long-term effects on disease progression and oral colonization. J Periodontol 76:749–759
American Academy of Periodontology (2000) Parameter on progressive periodontitis. J Periodontol 71:867–869
American Academy of Periodontology (2001) Treatment of plaque-induced gingivitis, chronic periodontitis, and other clinical conditions. J Periodontol 72:1790–1800
Beikler T, Prior K, Ehmke B, Flemmig TF (2004) Specific antibiotics in the treatment of periodontitis - A proposed strategy. J Periodontol 75:169–175
Beikler T, Karch H, Flemmig TF (2005) In German (Microbiologic diagnosis in periodontal therapy). Joint statement of the German Society of Periodontology (DGP) and the German Society of Dental, Oral, and Maxillofacial Medicine (DGZMK). Dtsch Zahnärztl Z 60:660–662
Krigar D-M, Kaltschmitt J, Krieger JK, Eickholz P (2007) Two subgingival plaque sampling strategies used with RNA-probes. J Periodontol 78:72–78
Baron F, Arndt R, Roßberg M, Schacher B, Wohlfeil M, Eickholz P (2008) In German (Prävalenz von Fusobacterium nucleatum und Prevotella intermedia in subgingivaler Plaque bei 2 Analysestrategien). Parodontologie 19:233–240
Beikler T, Schnitzer S, Abdeen G, Ehmke B, Eisenacher M, Flemmig TF (2006) Sampling strategy for intraoral detection of periodontal pathogenes before and following periodontal therapy. J Periodontol 77:1323–1332
Kim C-K, Choi S-H, Kim T-S, Kaltschmitt J, Eickholz P (2006) The infrabony defect and its determinants. J Periodontal Res 41:498–502
Fleiss JL (1981) Statistical methods for rates and proportions, 2nd edn. Wiley, New York, pp 223–225
van Winkelhoff AJ, Rodenburg JP, Goené RJ, Abbas EG, Winkel EG, de Graaff J (1989) Metronidazole plus amoxicillin in the treatment of Actinobacillus actinomycetemcomitans associated periodontitis. J Clin Periodontol 16:128–131
Dannewitz B, Pohl S, Eickholz P, Kim T-S (2007) Clinical and microbiological effects of a combined mechanic-antibiotic therapy in patients with Actinobacillus actinomycetemcomitans-associated periodontitis. Am J Dent 20:153–156
Eickholz P, Dannewitz B, Kim T-S (2005) Antibiotics in periodontal therapy. Perio 2:235–251
Ramich T, Schacher B, Scharf S, Röllke L, Arndt R, Eickholz P, Nickles K (2015) Subgingival plaque sampling after combined mechanical and antibiotic nonsurgical periodontal therapy. Clin Oral Investig 19:27–34
Eick S, Pfister W (2002) Comparison of microbial cultivation and a commercial PCR based method for detection of periodontopathogenetic species in subgingival plaque samples. J Clin Periodontol 29:638–644
Jervøe-Storm P-M, Koltzscher M, Falk W, Dörfler A, Jepsen S (2005) Comparison of culture and real-time PCR for detection and quantification of five putative periodontopathogenic bacteria in subgingival plaque samples. J Clin Periodontol 32:778–783
Mombelli A, McNabb H, Lang NP (1991) Black-pigmenting gram-negative bacteria in periodontal disease. I. Topographic distribution in the human dentition. J Periodontal Res 26:301–307
Jervøe-Storm PM, AlAhdab H, Koltzscher M, Fimmers R, Jepsen S (2010) Quantification of periodontal pathogens by paper point sampling from the coronal and apical aspect of periodontal lesions by real-time PCR. Clin Oral Investig 14:533–541
Jervøe-Storm P-M, AlAhdab H, Koltzscher M, Fimmers R, Jepsen S (2007) Comparison of curet and paper point sampling of subgingival bacteria as analyzed by real-time polymerase chain reaction. J Periodontol 78:909–917
Sixou M, Duffaut-Lagarrigue D, Lodter JP (1991) In French (A comparison between 4 different subgingival bacteriologic sampling technics). J Biol Buccale 19:16–21
Renvert S, Wikström M, Helmersson M, Dahlén G, Claffey N (1992) Comparative study of subgingival microbiological sampling techniques. J Periodontol 63:797–801
Hartroth B, Seyfahrt I, Conrads G (1999) Sampling of periodontal pathogens by paper points: evaluation of basic parameters. Oral Microbiol Immunol 14:326–330
Haffajee AD, Socransky SS (1992) Effect of sampling strategy on the false-negative rate for detection of selected subgingival species. Oral Microbiol Immunol 7:57–59
Casas A, Herrera D, Martin-Carnes J, Gonzáles I, O’Connor A, Sanz M (2007) Influence of sampling strategy on microbiologic results before and after periodontal therapy. J Periodontol 78:1103–1112
Wohlfeil M, Tabakci O, Arndt R, Eickholz P, Nickles K (2010) Detection rates of presumptive periodontal pathogens in subgingival plaque samples of untreated periodontitis using either four or six pooled samples. J Investig Clin Dent 1:126–132
Mombelli A, Casagni F, Madianos PN (2002) Can presence or absence of periodontal pathogens distinguish between subjects with chronic and aggressive periodontitis? A systematic review. J Clin Periodontol 29(suppl 3):10–21
Gatto MR, Montevecchi M, Paolucci M, Landini MP, Checchi L (2014) Prevalence of six periodontal pathogens in subgingival samples of Italian patients with chronic periodontitis. New Microbiol 37:517–524
Acknowledgments
This study was supported by Hain Lifescience GmbH, Nehren, Germany.
Conflict of interest
The authors report no conflicts of interest related to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nickles, K., Scharf, S., Röllke, L. et al. Detection of subgingival periodontal pathogens—comparison of two sampling strategies. Clin Oral Invest 20, 571–579 (2016). https://doi.org/10.1007/s00784-015-1530-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-015-1530-4