Abstract
Four novel ternary metal complexes of the type [M(Phen)(L1)2)] [phen = 1,10-phenanthroline, L1 = N-(2-hydroxyethyl)-N′-benzoylthiourea, M = Ni(II)(1), Co(II) (2), Cu(II) (3), Pd(II) (4)] were synthesized. The organic ligands and their corresponding organometallic complexes have been characterized using UV–vis absorption spectroscopy, element analysis, infrared radiation spectroscopy and fluorescence spectra. DNA binding and cleavage studies of these complexes were conducted in detail. In vitro DNA-binding properties were studied by electronic absorption spectra and fluorescence spectra methods. The results indicate that all of the ternary metal complexes can efficiently bind to DNA via intercalation mode. The DNA-binding constants for all ternary compounds are around 4 × 106 M−1. The binding propensity of the complexes to human serum albumin (HSA) was also investigated. Agarose gel electrophoresis study revealed that the metal complexes could cleave super-coiled pBR322 DNA to a nicked form in the absence of external agents. In vitro anti bacterial studies show that copper complex has weak antibacterial activities. Copper complex exhibits a better biological activity among all complexes. This study provides a new perspective and evaluation on the role and importance of the effect factors on the medicinal properties of benzoylthiourea compounds.
Graphical abstract
Synchronous fluorescence spectra of HSA (10 μM) as a function of concentration of the complexes 1–4.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thiourea and substituted thioureas [1] are extensively studied because they are a rich source of materials for the development of agrochemical and pharmaceutical products. Many of their important applications are plant-growth regulators [2, 3], herbicides [4, 5], insecticides [6], antifungal [7], antibacterial [3, 8], anti-HIV [9], anti tuberculosis and pharmacodynamics [10–12], protein inhibitors [13], etc. Recently, complexes containing substituted thiourea derivatives for Cu [14, 15], Co [15], Ni(II) [16, 17], Pd [18, 19], Pt [ [20–22] and Ru [23] attracted a wide attention to the researcher for their diverse range of antibacterial, antifungal, and anticancer activities. As an universal chelating ligand scaffold, 1,10-phenanthroline and its derivatives were widely reported having the ability to act on antifungal drugs [24, 25] even cytotoxic drugs [22, 26]. Metal complexes with phenanthroline ligand have strong ability of binding and cleaving DNA [27–29].
Nucleic acid is one of the main compositions of life. DNA regulates many biochemical processes occurring in the cellular system. Therefore, understanding and investigation of the interaction and mechanism of metal complexes binding with nucleic acid (DNA/tRNA) could be a more robust finding out of the origin of life on a molecular level and are particularly important for developing of new chemotherapy drugs in biological science. It is expected to design different ligands and metal complexes which could fit well in DNA binding, cleavage, as well as interactions with the certain protein which is the main cellular target for anticancer drugs. The interaction of drug with human serum albumin (HSA) and bovine serum albumin (BSA) will provide important information on the absorption, distribution, activity and toxicity of a specific drug. In our previous works, we prepared a series of N-benzoyl-N′-arylthiourea derivatives and their binary metal (Cu, Pt, Pd) complexes. The antibacterial properties of the ligands and complexes against Gram-positive and Gram-negative bacteria were investigated [30, 31]. The incorporation of transition metal center into such structures can not only be used to impart cationic charge, but also to confer new properties on these molecules [22]. Some acylthiourea binary metal complexes frequently were synthesized. They usually exhibit cytotoxicity against tumour cells, and are more cytotoxic than the free ligands [13, 14, 20, 22, 23]. However, the practical result for ternary metal complexes involving benzoylthiourea derivatives has rarely been reported.
To further investigate the effect on the medicinal and biological properties of the ternary metal (Co, Ni, Cu, Pd) complexes with benzoylthiourea series, we synthesized four kinds of [M(Phen)(L1)2] complexes using N-(2-hydroxyethyl)-N′-benzoylthiourea and 1,10-phenanthroline as the ligands. Four different metals were introduced to contrast and compare the difference of their biological activity. In addition, the interaction of these complexes with HSA, cleavage activity with pBR322 plasmid DNA by agarose gel electrophoresis method. DNA-binding profile and antibacterial activity were studied in vitro. The results demonstrated that all of the ternary complexes could intercalate with Calf-Thymus DNA (CT-DNA) and bind to HSA in different degree. The ternary complex of copper exhibits a better biological activity out of four metal compounds synthesized. The aim of this study is to provide a new perspective and evaluation on role and importance of the effect factors on the medicinal properties of benzoylthiourea and transition metal. The incorporation of thioureas derivatives or phen as ancillary ligands in mixed compounds is a promising strategy to design new anticancer metal compounds with intercalating properties.
Materials and methods
Chemicals and instrumentation
All reagents used were of analytical grade unless otherwise stated. Co(OO-CCH3)2·4H2O and Ni(OO-CCH3)2·4H2O were obtained from chemical industry company of Tianjin Kaixin. Cu(OO-CCH3)2·H2O was purchased from chemical industry company of Shanghai Jianxin. PdCl2 and Phen were purchased from Damao chemical company of Tianjin. Calf-Thymus DNA (an average size of 100 to 2000 base pairs, 2 mg/ml, Shipping Condition: Dry ice, A 260/A 280 > 1.9,) was purchased from Solarbio Life Sciences. HSA was obtained from Rongsheng Pharmacy Company of Chengdu (China). Super-coiled pBR322 DNA in which open circular relaxed form (Form II) was major components was purchased from Dingguo Changsheng Biotechnology Company of Beijing (China). Agarose (molecular biology grade) and ethidium bromide were obtained from Zhongqin chemical company of Shanghai (China). 10 mM Tris–HCl/10 mM NaCl buffer solution being used for DNA and HSA binding or pBR322 DNA cleavage (1 μg/μL, 4631 bp) was prepared using ultrapure water. Tris–acetate–EDTA (TAE) buffer was used for DNA cleavage study. All of the absorption spectra were performed using a UV–Vis Spectrophotometer (T6, Beijing Puxi Instrument Co., Beijing, China) with 1.00 cm quartz cuvettes. The fluorescence spectra of the compounds were recorded in solution on a Shanghai F-280 fluorescence spectrophotometer (F-280, Shanghai Analytical Instrument Factory, Shanghai, China). Vibration spectra were performed on FT–IR–double–beam model IR spectrophotometer (Tianjin Guangdong Science and Technology Development Limited Company, Tianjin, China) with samples prepared as KBr pellets.
Synthesis of N-(2-hydroxyethyl)-N′-benzoylthiourea
The benzoylthiourea ligands were prepared according to the reported methods [30–32]. Benzoyl isothiocyanate was obtained by the reaction of benzoyl chloride (1.410 g, 10 mmol) and ammonium thiocyanate (1.140 g, 15 mmol) in CH2Cl2 (20 mL) solution using polyethylene glycol-400 (0.180 g) as solid–liquid phase transfer catalyst. The benzoyl isothiocyanate was then treated with ethanolamine (0.610 g, 10 mmol) to give the objective compound. The final solid was separated from the reaction mixtures by filtration, washed with CH2Cl2, and followed by recrystallisation from ethanol then dried at 40 °C.
N-(2-hydroxyethyl)-N′-benzoylthiourea (L 1 ): White needle crystal, m.p., 124–126 °C, and UV–Vis (acetonitrile in solution): λ max (nm), 242, 280. Fluorescence spectroscopy (DMSO in solution): λ ex (nm), 303, λ em (nm), 337. FT-IR (KBr): ν (cm−1), 1670 (C=O), 1270 (C=S), 3332, 3232(N–H).
The ligand of 1,10-phenanthroline (phen)
For white needle crystal, UV–Vis (acetonitrile in solution): λ max (nm), 230, 264. Fluorescence spectroscopy (DMSO in solution): λ ex (nm), 302, λ em (nm), 337. FT-IR (KBr): ν (cm−1), 1504 (ring), 1090 (C–H), 736 (C–H).
Synthesis of the complexes
Ternary complex of Ni, Co, and Cu (complex 1–3)
1 mmol M(Ac)2 and 1 mmol phen were dissolved in 20 mL 50 % ethanol solution forming colored solution, the solution was added to benzoylthiourea (2 mmol) 95 % ethanol solution. The reaction mixture was heated at 40 °C with constant stirring. Then, the resulting precipitate was separated by filtration washing several times with water and ethanol to afford pure and stable compounds.
[Ni(phen)(L1)2] (1): Purple powder, Anal. calcd for C32H32N6S2O4Ni: C, 55.90; H, 4.70; N, 12.23; S, 9.33. Found: C, 55.49; H, 3.12; N, 12.66; S, 9.01 (%). UV–Vis (acetonitrile in solution): λ max (nm), 228, 270. Fluorescence spectroscopy (DMSO in solution): λ ex (nm), 376, λ em (nm), 410. FT-IR (KBr): ν (cm−1), 1653 (C=O), 1221 (C=S), 3426 (N–H).
[Co(phen)(L1)2] (2): Red Powder, Anal. calcd for C32H32N6S2O4Co: C, 55.51; H, 4.70; N, 12.22; S, 9.33. Found: C, 56.67; H, 2.92; N, 13.21; S, 9.01 (%). UV–Vis (acetonitrile in solution): λ max (nm), 226,268. Fluorescence spectroscopy (DMSO in solution): λ ex (nm), 374, λ em (nm), 408. FT-IR (KBr): ν (cm-1), 1627 (C=O), 1222 (C=S), 3428(N–H).
[Cu(phen)(L1)2] (3): Snow blue powder, Anal. calcd for C32H32N6S2O4Cu: C, 55.88; H, 4.67; N, 12.14; S, 9.26. Found: C, 55.13; H, 3.55; N, 11.46; S, 9.06 (%). UV–Vis (acetonitrile in solution): λ max (nm), 236, 264. Fluorescence spectroscopy (DMSO in solution): λ ex (nm), 376, λ em (nm), 409. FT-IR (KBr): ν (cm−1), 1596 (C=O), 1266 (C=S), 3436 (N–H).
Ternary complex of Pd (complex 4)
The Pd(II) complex was prepared according to the reported procedures [33]. The complex [Pd(phen)Cl2] was obtained by the reaction of phen (1 mmol) with K2[PdCl4] (1 mmol) in methanol solution under reflux about 1 h. The precipitated crystalline powders were recovered by filtration and dried under vacuum for 2 h. The resulting complex was suspended again in a water/methanol solution; the respective benzoylthiourea (1 mmol, L1) was then added to the suspension under reflux. After 1 h, clear yellow to orange solution was obtained. The solution was filtrated and the filtrates were kept for 3–5 days at room temperature for crystallization. As a result, yellow–red crystals were obtained.
[Pd(phen)(L1)2] (4): Orange solid, Anal. calcd for C32H32N6S2O4Pd: C, 52.27; H, 4.40; N, 11.43; S, 8.72. Found: C, 52.11; H, 3.63; N, 11.06; S, 8.46 (%). UV–Vis (methanol in solution): λ max (nm), 236, 276. FT-IR (KBr): ν (cm−1), 1578 (C=O), 1260 (C=S), 3400 (N–H). Fluorescence spectroscopy (methanol in solution): λ ex = 277 nm, λ em = 577 nm.
DNA-binding activity
The DNA-binding experiments were performed at room temperature. M(II) complex-DNA solutions were allowed to incubate for 10 min before each of measurements was carried out. Absorption spectral traces and fluorescence methods were used to determine the binding modes of the complexes to Calf-Thymus DNA.
For fluorescence-quenching experiments, DNA was pre-treated with ethidium bromide (EB) for 30 min. The M(II) complex samples were added to this mixture, and then, the emission intensity was measured. The emission intensity was observed between 550 and 750 nm when the samples were excited at 522 nm.
DNA cleavage experiments
For the gel electrophoresis study, super-coiled pBR322 DNA (1.0 μg/μL) was treated with the M(II) complex in Tris–HCl buffer, pH 7.42. The solution was incubated for 1 h in a dark chamber at 37 °C. The samples were electrophoresed 0.5 h at 80 V on a 0.9 % agarose gel in TAE buffer. The gel was stained with 0.5 μg EB before the electrophoresis. Bands were visualized by UV light and photographed. The extent of cleavage was measured using the UVITEC Gel Documentation System [34].
HSA-binding experiments
On additional of metal complex solution to 10 μM HSA solution, the solution was determined from absorption spectra at 278 nm using Tris–HCl buffer solution as reference. The HSA-binding study was also carried out with excitation wavelength at 293 nm and emission wavelength at 330 nm using a fluorescence spectrophotometer. The excitation and emission slit widths, sensitivity, and scan rates were constantly maintained for all the experiments.
Synchronous fluorescence spectra were recorded using the same concentration of HSA and the complexes as mentioned above with two different ∆λ (difference between the excitation and emission wavelengths of HSA) values, such as 15 and 60 nm.
Antifungal assay
The antibacterial test was done in vitro against one Gram-negative bacteria standard strains (Escherichia coli) and one Gram-positive standard bacteria strains (Staphylococcus aureus) using the agar well diffusion method [35, 36]. Each M(II) complex and benzoylthiourea (1 × 10−3 M) in dimethyl sulphoxide(DMSO)or methanol solution was prepared for testing against spore germination of each fungus. Filter paper discs (5 mm) were prepared using Whatman filter paper No.1 (sterilized in an autoclave) which was saturated with 10 μL of the compounds solution. The fungal culture plates were incubated at 37 ± 0.2 °C for 24 h to make sure that no contamination would occur in the dish. Activity was determined by measuring the diameter of complete inhibition zone (in mm).
Results and discussion
Characterization of ligands and metal complexes
IR spectroscopy
In the infrared spectra, the bands of the ring-stretching frequencies [ν (C=C) and ν (C=N)] at 1504 and 1422 cm−1 of free phenanthroline were shifted to higher frequencies upon complexation. The C–H band at 852 and 736 cm−1 in the spectra of phenanthroline was also shifted upon metal complexation. The shifts can be explained by the fact that each of the two nitrogen atoms of phenanthroline ligand offers a pair of electrons to the central metal atom forming a coordinate covalent bond [37, 38]. The bands at 3332 and 3232 cm−1 could be assigned to the N–H stretches of the benzoylthiourea, while the bands were also shifted to higher frequencies and turned into a broad peak in the metal complexes due to intramolecular hydrogen bonding in benzoylthiourea. The bands observed at 1222 and 1670 cm−1 in L1 were assigned to C=S and C=O stretching vibrations, respectively. The C=S and C=O were shifted to lower frequencies about 4–49 and 17–92 cm−1 in the complexes. The carbonyl O atom in L1 lose coordination capability due to intramolecular hydrogen bonding [39], so L1 ligand behaves like a monodentate thiourea ligand [32].
UV–Vis absorption spectroscopy
The free phen ligand exhibits absorption bands at 230 and 264 nm, whilst two absorption bands in the range 235–290 nm are observed for the free benzoylthiourea ligand (L1). The UV–vis spectra of the complexes (1–4) in methanol or acetonitrile present one to two absorption bands located in the UV range, which can be assigned to intraligand n–π* and π–π* transitions (LC) perturbed by complexation to the M(II) metal. The moderately intense absorptions around 270 nm for all prepared complexes 1–4 can be attributed to spin-allowed metal-to-ligand charge transfer MLCT(dM–π*) transition in analogy to semblable metal complexes containing phen ligand [40].
Fluorescence spectroscopy
All the complexes show strong luminescence properties both in DMSO and methanol solution at room temperature. The absorption spectra are closely matched with the excitation spectra. All metal complexes show two emission bands in the 390–500 nm range except for the emission band of Pd(II) complex in the 500–650 nm range. The first one has the same position around 409 nm for complexes 1–3, but in respect to the position of the complex 4 is around 539 nm. For complexes 1–3, the first band intensity is higher than the second one located at 428 nm; whilst for complex 4, this band has higher intensity compared with the second emission band at 577 nm. Moreover, the emission band is a clear red shift for complexes and the fluorescence intensity has an apparent change compared to the ligands. The results show that the electron of excited state goes back to ground state resulting in the change of fluorescence intensity upon complexation for ligands and metal ions.
DNA-binding studies
Electronic absorption study
Titration with UV spectrophotometry is well suited for investigating the binding mode of DNA with metal complexes. The extent of the hypochromism or hyperchromism in the metal-to-ligand charge transfer (MLCT) band is in accordance with the strength of intercalative interaction [41].
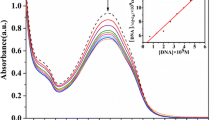
Upon addition of increasing concentration of DNA to the fixed concentration of complex (1-4), a concomitant increase in the absorption intensity (hyperchromism) was observed, as shown in Fig. 1. In the presence of increasing concentrations of CT-DNA, a hyperchromism effect is apparent. Hypochromism possibly results from the interaction between the electronic states of the intercalating chromophore and that of the DNA base [42]. The association reaction between the ternary complexes and CT-DNA at low concentration level is of intercalation mode which is most likely through the mode involving a stacking interaction between the aromatic chromophore and the base pairs of DNA [43].
a–d UV–Vis spectra of the complexes 1–4 in Tris–HCl buffer upon addition of Calf-Thymus DNA (a–i), [complex] = 2.0 × 10−5 M; a = 0.00, b = 0.17, c = 0.33, d = 0.50, e = 0.66, f = 0.82, g = 0.98, h = 1.14, and i = 1.30 μM. Arrow shows the absorbance changes with increasing DNA concentration. Inset plots of [DNA]/(ε a − ε f) vs. [DNA] for the titration of complexes 1–4 with ct-DNA
The DNA-binding affinities for the interaction of the complex with DNA are compared quantitatively by obtaining the intrinsic binding constant K b using the following the following equation [44]:
where [DNA] is the concentration of DNA in base pairs, ε a, ε f, and ε b correspond to the apparent absorption coefficient A obs/[complex], the extinction coefficient for the free metal complex and the extinction coefficient for the metal complex in the fully bound form, respectively. In plots of [DNA]/(ε a − ε f) versus [DNA], K b is afforded by the ratio of slope to the intercept. The K b values were listed at Table 1. All K b values for the complexes are higher than complexes like [(Ru(phen)2(bpy)](PF6)2 (K b = 0.36 × 104 M−1), tricationic Co(III) complexes with asymmetric ligand, [Co (phen)2(pdta)]3+(K b = 2.8 × 104 M−1), and [Co(bpy)2(CNOIP)]3+(K b = 5 × 104 M−1) [43]. Obviously, the rigid structure of ternary complexes would lead to much stronger interaction with DNA. While the result suggests that different metals do not influence the intercalating ability of these complexes.
Fluorescence spectral studies
On addition of increasing concentration of CT-DNA (0–1.44 μM) to fixed amount of complexes 1–3, fluorescence emission intensity increased remarkably with slight change in the shape and position of emission bands (Fig. 2). The result indicates that complexes 1–3 can strongly interact with DNA and be protected efficiently from the hydrophobic environment inside the DNA helix. Since DNA being hydrophobic macromolecule binds to complex, the emission spectrum of complexes is not quenched. The accessibility of solvent molecules to the duplex is reduced and the complex mobility is limited at the binding site, which leads to the decrease in the vibrational modes of relaxation [45]. However, the decrease of fluorescence intensity for complex 4 is most likely attributed to energy or electron transfer from the guanine base of DNA to the MLCT of complex [46].
a–d Fluorescence emission spectra of the complex 1–4 in DMSO or methanol solution upon addition of Calf-Thymus DNA (a–h), [complex] = 4.0 × 10−5 M; a = 0.00, b = 0.17, c = 0.33, d = 0.50, e = 0.66, f = 0.82, g = 0.98, h = 1.14 μM, i = 1.30 μM, and j = 1.44 μM. Arrow shows the emission intensity changes with increasing DNA concentration. Inset Stern–Volmer plot of the fluorescence titration data of the complexes 1–4
EB displacement assay
To further investigate the binding mode of complexes 1–4 with DNA, competitive-binding experiment using ethidium bromide (EB) as a probe was implemented. The present M(II) complexes can quench the EB emission by either displacing EB of DNA-EB system or involving in partial intercalation of planar ring at higher concentrations [47]. The fluorescence intensity decreases with the increment amount of complexes (1–4), as shown in Fig. 3.
The results suggest that complexes might bind to CT-DNA through intercalated interaction to a large extent, which is the most possible interaction mode to CT-DNA. The Stern–Volmer quenching constant K q for each complex was calculated using the equation given as F 0 /F = 1 + K q [Q]. Here, F 0 and F are the fluorescence intensity in the absence and presence of complexes, respectively, K q is a linear Stern–Volmer quenching constant, and [Q] is the total concentration of a complex to that of DNA. The value of K q is given by the ratio of slope to intercept in a plot of F 0/F versus [Q] (Fig. 4).
The quenching constant values are listed in Table 1. To quantify the displacement, the concentration of the complex at which EB fluorescence decreases by 50 % (assumed to be 50 % displacement of EB) is calculated [48]. From a plot of the observed intensities against complex concentration, the values of apparent DNA-binding constant (K app) were calculated using the equation K EB[EB] = K app[complex], where K EB (4.94 × 105 M−1) is the DNA-binding constant of EB, [EB] is the concentration of EB (1.25 μM), and [complex] is the concentration of the complex used to obtain 50 % reduction in fluorescence intensity of EB. The higher K q values reveal that the ternary complexes have stronger ability of displacing EB from DNA-EB system. The capability of the complexes to quench the fluorescence in EB displacement assay is in accordance with the UV–Vis titration spectroscopic studies, which indicate their intercalation in DNA.
DNA cleavage studies
To assess the DNA cleavage ability of the complexes, super-coiled plasmid pBR322 DNA was cultured with complexes 1–4 in Tris–HCl buffer solution at pH 7.42 for 1 h in the absence of any external agents. In the absence of the complex, the DNA remained in its super-coiled form (Form I). Upon addition of complex when subjected to agarose gel electrophoresis, an open circular relaxed form (Form II) was formed and the linear form (Form III) was found upon further cleavage. When electrophoresis was performed on the reaction mixture, the compact Form I migrated relatively faster, while the nicked Form II migrated slowly. The linearized form (Form III) migrated between Forms I and II [49]. From Fig. 5, all the present complexes converted open circular relaxed form (Form II) into linearized form (Form III) to almost the same extent. It was found that all the metal complexes could effectively cleave the super-coiled DNA. The nuclease activity of the Cu(II) complex shows the highest ability of cleaving DNA and with the order Cu > Ni > Pd > Co. It can be seen that ternary complexes of copper show efficient chemical nuclease activity and are good DNA intercalative binders [14] and have the potential for applications in antibacterial agents.
Gel electrophoresis diagram showing the cleavage of super-coiled pBR322 DNA (1 μg/μL in base pair) by complexes 1–4 (100 μM) in DMSO or methanol/Tris–HCl buffer at pH 7.42 and 37 °C with an incubation time 1 h in the absence of any reducing/oxidative agents. Lane 1 DNA, lane 2 DNA + Ni, lane 3, DNA + Co, lane 4 DNA + Cu, lane 5 DNA + Pd
HSA-binding studies
Most of the total blood plasma protein is constituted of serum albumin. The protein plays a vital role in the drug transport (metal complexes) and drug metabolism. The binding affinity of complex towards HSA was also investigated to understand the carrier role in body system.
Electronic absorption spectroscopy
Upon addition of complexes 1–4 to HSA of constant concentration (10 μM), a gradual increase in absorption intensity and hyperchromism of the intraligand band at 278 nm was observed, as shown in Fig. 6.
a–d Electronic absorption titration curves of HSA (10 μM) in the absence and presence of the M(II) complex in Tris–HCl buffer (pH 7.20). The arrow shows the absorbance changes with increasing the concentration of the complex (a–j), a = 0, b = 0.1, c = 0.3, d = 0.5, e = 0.7, f = 1.0, g = 1.4, h = 1.9, i = 2.3, and j = 2.7 μM. Inset plot of 1/(A − A 0) vs. 1/[M(II) complex
The results indicate that the complexes 1–4 can interact with HSA by non-covalent interaction possibly bind to HSA by the modes of hydrophobic interaction and electrostatic binding. Electrostatic attraction can be facilitated by hydrogen-bond formation, which results in the slight change in the absorption profile deriving from the effect of the polar solvent (water) and perturbations in the microenvironment of the polypeptide backbone of the HSA [50]. The plots of 1/(A − A 0) vs. 1/[M(II) complex] are linear lines (Fig. 6), and the binding constant (K b) can be obtained from the ratio of the intercept to slope. The binding constant (K b) of 5.1 × 104, 4.4 × 105, 5.7 × 105, and 1.3 × 105 M−1 for Ni(II), Co(II), Cu(II) and Pd(II) complex with HSA were calculated, respectively. The values of K b show that HSA can be considered as a good carrier for transfer of these complexes in vivo.
Fluorescence-quenching studies with HSA
Binding to the protein may lead to the loss or enhancement of the biological properties of the original compounds, or provide paths for their transportation. Fluorescence-quenching analyses are usually employed to analyze the interaction of chemical compounds with HSA. A solution of HSA (10 μM) was titrated with various concentration of the complexes (0.1–0.6/0.7 μM). The fluorescence spectrum of tryptophan was recorded in the range from 290 to 500 nm upon excitation at 293 nm. The effects of complexes (1–4) on the fluorescence spectrum of HSA are shown in Fig. 7.
a–d Intrinsic tryptophan fluorescence spectra of HSA (10 μM) in the presence of complex 1–4 (a–g/h), a = 0, b = 0.1, c = 0.2, d = 0.3, e = 0.4, f = 0.5, g = 0.6, and h = 0.7 μM. Arrows indicate the effect of increasing concentration of complexes on the tryptophan fluorescence emission of HSA. The inset shows the linear fit of F 0/F vs. [complex]
Upon the addition of the M(II) complexes to the solution of HSA, fluorescence intensity at 330 nm decreases with the initial intensity of HSA, accompanied by a hypsochromatic shift of 6, 9, 4, and 5 nm for complexes 1, 2, 3, and 4. The observed hypochromicity with blue shift reveals that the complexes could interact hydrophobically with the HSA protein and the efficient interaction between metal complex and HSA results in the change in its secondary structure (affecting the tryptophan residues of HSA).
The Stern–Volmer and Scatchard graph is used to study the interaction of a quencher with serum albumins. According to the Stern–Volmer quenching the following equation [51]:
where F 0 is the initial tryptophan fluorescence intensity of HSA, F is the tryptophan fluorescence intensity of HSA after the addition of the quencher, k q is the quenching rate constants of HSA, K SV is the dynamic quenching constant, τ o is the average lifetime of HSA without the quencher, [Q] is the concentration of the quencher, K SV = k q τ o, and taking as fluorescence lifetime (τ o) of tryptophan in HSA at around 10−8 s, and the dynamic quenching constant (K SV, M−1) can be calculated using the plot of F 0/F versus [Q] (Fig. 8).
If it is assumed that the binding of complexes with HSA occurs at equilibrium, the equilibrium binding constant can be analyzed according to the Scatchard equation [52]:
where K b is the binding constant of the compound with HSA and n is the number of binding sites. From the plot of log [(F 0 − F)/F] versus log [Q] (Fig. 9), the number of binding sites (n) and the binding constant (K b) values were obtained. The calculated K SV, K b, and n values were given in Table 2. The value of n was around 1.2 for all of the complexes. Notably, Pd(II) complex exhibited a better interaction effect with HSA than other complexes.
Characteristics of synchronous fluorescence spectra
Synchronous fluorescence spectroscopy offers information on the molecular microenvironment, particularly in the vicinity of the fluorophore functional groups. The presence of tyrosine, tryptophan, and phenylalanine residues might have an influence on the fluorescence of HSA. The difference between excitation and emission wavelengths (Δλ) reflects the nature of chromosphere [53]. The large (~60 nm) and the small (~15 nm) Δλ values are characteristic of tryptophan and tyrosine residues, respectively.
Synchronous fluorescence spectra of HSA were recorded at both 15 and 60 nm Δλ with the addition of the M(II) complexes (1–4) in various concentration to understand the structural change occurred in HSA (Figs. 10, 11).
With increasing the concentration of the complexes, the intensity of emission corresponding to tyrosine (at 293 nm) was found to decrease in the magnitude of 58.7, 63.5, 53.2, and 41.5 % for complexes 1–4 accompanied with slightly red shift in emission wavelength. The tryptophan fluorescence emission presented apparent decrease in the intensity (at 288 nm) of about 38.8, 40.7, 43.4, and 31.6 % for complexes 1, 2, 3, and 4. The position of the emission band had a slight shift. The results indicate that the complexes could affect the microenvironments of both tyrosine and tryptophan during the binding process, while the effect was more towards tyrosine than tryptophan [54].
Antibacterial activities
All synthesized compounds were screened for antibacterial activity against bacterial strains by the inhibition zone method with agar diffusion. The biological activity for the complexes was defined by measuring the diameter of the inhibition area surrounding each bore in millimeters. The experiments were repeated three parallel times. The values of inhibition zones of the all compounds were reported as means of at least three determinations and were represented in Table 3.
The antibacterial activities of the complexes and ligand were determined against Gram-positive and Gram-negative pathogenic organisms. Methicillin [R (≤9 mm), I (10–13 mm), S (≥14 mm)] and trospectomycin (I = 10–16 mm, S = 15–20 mm) were used as the standard control. The results show that only copper complexes have nearly moderate activity against the tested bacterial strains. All complexes have slightly higher activity than their ligands against the same microorganism. The antibacterial activities of copper complex shows more activity against the tested bacteria than other metals and with the order Cu > Pd > Co > Ni. Weak antibacterial activity of Pd, Co, and Ni complexes may attribute to the poor solubility in solvent. It should be emphasize that the antibacterial study is very preliminary, since it was based on only two bacteria.
Conclusion
N, N′-disubstituted thiourea derivatives and 1,10-phenanthroline complexes still receive much attention due to synthetic convenience and excellent biological activities. O-, S-, and N-donor atoms and thione, thiol (–N–CS–N–) groups in the ligands stretch their coordination ability and popularity towards a variety of metal ions [14]. Four novel ternary metal complexes of N-benzoylthiourea and 1,10-phenanthroline derivatives, [Ni(phen)(L1)2], [Co(phen)(L1)2], [Cu(phen)(L1)2], and [Pd(phen)(L1)2], have been synthesized and characterized by their conventional physical and chemical analyses. UV–Vis and fluorescence spectral studies indicate that all these complexes exhibit efficient and strong binding abilities with DNA and HSA. The experiments of fluorescence quenching further reveal the strong interaction of the complexes with HSA protein. Meanwhile, all ternary complexes can cleavage DNA efficiently. Among the complexes examined, copper (II) complex exhibits the strongest DNA binding and cleavage ability. Although this study is preliminary, they provide the theoretical basis and experimental data for the study of the potential drug with nucleic acid as the target based on disubstituted thioureas. We hope to inspire the role and importance of N, N′-disubstituted thiourea compounds and further work on their complexes for biomedical applications.
Abbreviations
- HSA:
-
Human serum albumin
- DNA:
-
Deoxyribose nucleic acid
- CT-DNA:
-
Calf-thymus DNA
- DMSO:
-
Dimethyl sulphoxide
- BSA:
-
Bovine serum albumin
- Tris-HCl:
-
Tris(hydroxymethyl)aminomethane
- EB:
-
Ethidium bromide
References
Aly AA, Ahmed EK, El-Mokadem KM, Hegazy ME-AF (2007) J Sulfur Chem 28:73–93
Lin Q, Yao H, Wei T, Zhang Y (2009) Indian J Chem 48B:124–127
Zhao MM, Dong XY, Yang YH, Li G, Zhang YJ (2014) Asian J Chem 26:237–240
Xue S, Ke S, Duan L (2004) Chin J Org Chem 24:227–230
Peng H, He H (2007) Chin J Org Chem 27:502–506
Xu Z, Liu B, Dong H, Wang M (2014) Chin J Org Chem 34:2517–2522
del Campo R, Criado JJ, Garcı́a E, Hermosa MAR, Jiménez-Sánchez A, Manzano JL et al (2002) J Inorg Biochem 89:74–82
Kurt G, Sevgi F, Mercimek B (2009) Chem Pap 63:548–553
Stefanska J, Szulczyk D, Koziol AE, Miroslaw B, Kedzierska E, Fidecka S et al (2012) Eur J Med Chem 55:205–213
Tahir S, Badshah A, Hussain RA, Tahir MN, Tabassum S, Patujo JA et al (2015) J Mol Struct 1099:215–225
Wang M-J, Nan X, Feng G, Yu H-T, Hu G-F, Liu Y-Q (2014) Ind Crops Prod 55:11–18
Maruyama T, Seki N, Onda K, Suzuki T, Kawazoe S, Hayakawa M et al (2009) Biorg Med Chem 17:5510–5519
Zeng Y-Q, Cao R-Y, Yang J-L, Li X-Z, Li S, Zhong W (2016) Eur J Med Chem 119:83–95
Chetana PR, Srinatha BS, Somashekar MN, Policegoudra RS (2016) J Mol Struct 1106:352–365
Singh A, Bharty MK, Bharati P, Bharti A, Singh S, Singh NK (2015) Polyhedron 85:918–925
Selvakumaran N, Pratheepkumar A, Ng SW, Tiekink ERT, Karvembu R (2013) Inorg Chim Acta 404:82–87
Rauf MK, Yaseen S, Badshah A, Zaib S, Arshad R, Imtiaz-Ud-Din, et al (2015) J Biol Inorg Chem 20(3):541–554
Plutín AM, Mocelo R, Alvarez A, Ramos R, Castellano EE, Cominetti MR et al (2014) J Inorg Biochem 134:76–82
El-Ayaan U (2011) J Mol Struct 998:11–19
Plutín AM, Alvarez A, Mocelo R, Ramos R, Castellano EE, da Silva MM et al (2016) Inorg Chem Commun 63:74–80
Correa RS, Oliveira KM, Pérez H, Plutín AM, Ramos R, Mocelo R, et al. (2015) Arab J Chem. doi:10.1016/j.arabjc.2015.10.006
Barra CV, Rocha FV, Morel L, Gautier A, Garrido SS, Mauro AE et al (2016) Inorg Chim Acta 446:54–60
Correa RS, de Oliveira KM, Delolo FG, Alvarez A, Mocelo R, Plutin AM et al (2015) J Inorg Biochem 150:63–71
Sudhamani CN, Bhojya Naik HS, Sangeetha Gowda KR, Giridhar M, Girija D, Prashanth Kumar PN (2015) Spectrochim Acta A 138:780–788
Kashanian S, Khodaei MM, Roshanfekr H, Peyman H (2013) Spectrochim Acta A 114:642–649
Gurumoorthy P, Mahendiran D, Prabhu D, Arulvasu C, Rahiman AK (2015) J Mol Struct 1080:88–98
Ganeshpandian M, Ramakrishnan S, Palaniandavar M, Suresh E, Riyasdeen A, Akbarsha MA (2014) J Inorg Biochem 140:202–212
Thamilarasan V, Jayamani A, Sengottuvelan N (2015) Eur J Med Chem 89:266–278
Kannan D, Arumugham M (2013) Int J Inorg Bioinorg Chem 3:8–15
Peng B, Li T, Zhang Z, Shen Y, Zhou M, Mo Z (2014) Chem Reagents 36:205–261
Peng B, Lin B, Zhang Z, Chen P, Ma S (2012) J Northwest Norm Univ Nat Sci 48:51–56
Koch KR, Sacht C, Bourne S (1995) Inorg Chim Acta 232:109–115
Rotondo A, Barresi S, Cusumano M, Rotondo E (2012) Polyhedron 45:23–29
Rao R, Patra AK, Chetana PR (2008) Polyhedron 27:1343–1352
Srishailam A, Gabra NM, Kumar YP, Reddy KL, Devi CS, Anil Kumar D et al (2014) J Photochem Photobiol, B 141:47–58
Devi J, Batra N (2015) Spectrochim Acta A 135:710–719
Reddy PR, Shilpa A, Raju N, Raghavaiah P (2011) J Inorg Biochem 105:1603–1612
Leela DS, Ushaiah B, Anupama G, Sunitha M, Kumari CG (2015) J Fluoresc 25:185–197
Zhang Y, Xian L, Wei T, Cai L (2003) Acta Cryst E E59:o817–o819
Cîrcu V, Ilie M, Iliş M, Dumitraşcu F, Neagoe I, Păsculescu S (2009) Polyhedron 28:3739–3746
Patel MN, Gandhi DS, Parmar PA, Joshi HN (2012) J Coord Chem 65:1926–1936
Barcelo F, Barcelo I, Gavilanes F, Ferragut JA, Yanovich S, Gonzales-Ros JM (1986) Biochim Biophys Acta 884:172–181
Liu H, Li L, Guo Q, Dong J, Li J (2013) Transition Met Chem 38:441–448
Zhai S, Guo Q, Dong J, Xu T, Li L (2014) Transition Met Chem 39:271–280
Tabassum S, Zaki M, Afzal M, Arjmand F (2014) Eur J Med Chem 74:509–523
Tabassum S, Ahmad M, Afzal M, Zaki M, Bharadwaj PK (2014) J Photochem Photobiol, B 140:321–331
Li R, Lu J, Li D, Cheng S, Dou J (2014) Transition Met Chem 39:507–517
Borowska J, Sierant M, Sochacka E, Sanna D, Lodyga-Chruscinska E (2015) J Biol Inorg Chem 20:989–1004
Moosun SB, Jhaumeer-Laulloo S, Hosten EC, Gerber TIA, Bhowon MG (2015) Transition Met Chem 40:445–458
Tabassum S, Zaki M, Ahmad M, Afzal M, Srivastav S, Srikrishna S et al (2014) Eur J Med Chem 83:141–154
Lakowicz JR, Weber G (1973) Biochemistry-US 12:4161–4170
Anjomshoa M, Torkzadeh-Mahani M (2015) Spectrochim Acta A 150:390–402
Miller JN (1979) Proc Anal Div Chem 16:203–208
Selvakumaran N, Bhuvanesh NSP, Endo A, Karvembu R (2014) Polyhedron 75:95–109
Acknowledgments
The authors would like to thank the National Natural Science Foundation of China (21174114, 21167015), the Innovative Team of Ministry of Education of China (IRT15R56), and “Integration and demonstration of the key technology of industrial capacity promotion for Lanzhou Lily” (2015 technological innovation special fiscal funds of Gansu province) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peng, B., Gao, Z., Li, X. et al. DNA binding, DNA cleavage and HSA interaction of several metal complexes containing N-(2-hydroxyethyl)-N′-benzoylthiourea and 1,10-phenanthroline ligands. J Biol Inorg Chem 21, 903–916 (2016). https://doi.org/10.1007/s00775-016-1388-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-016-1388-1