Abstract
The protean chemical properties of the toxic metal mercury (Hg) have made it attractive in diverse applications since antiquity. However, growing public concern has led to an international agreement to decrease its impact on health and the environment. During a recent proteomics study of acute Hg exposure in E. coli, we also examined the effects of inorganic and organic Hg compounds on thiol and metal homeostases. On brief exposure, lower concentrations of divalent inorganic mercury Hg(II) blocked bulk cellular thiols and protein-associated thiols more completely than higher concentrations of monovalent organomercurials, phenylmercuric acetate (PMA) and merthiolate (MT). Cells bound Hg(II) and PMA in excess of their available thiol ligands; X-ray absorption spectroscopy indicated nitrogens as likely additional ligands. The mercurials released protein-bound iron (Fe) more effectively than common organic oxidants and all disturbed the Na+/K+ electrolyte balance, but none provoked efflux of six essential transition metals including Fe. PMA and MT made stable cysteine monothiol adducts in many Fe-binding proteins, but stable Hg(II) adducts were only seen in CysXxx(n)Cys peptides. We conclude that on acute exposure: (a) the distinct effects of mercurials on thiol and Fe homeostases reflected their different uptake and valences; (b) their similar effects on essential metal and electrolyte homeostases reflected the energy dependence of these processes; and (c) peptide phenylmercury-adducts were more stable or detectable in mass spectrometry than Hg(II)-adducts. These first in vivo observations in a well-defined model organism reveal differences upon acute exposure to inorganic and organic mercurials that may underlie their distinct toxicology.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The toxic metallic element mercury occurs naturally as the insoluble HgS ore (cinnabar), as soluble inorganic complexes of Hg+2, Hg+1, or (Hg2)2+ with counterions such as acetate, nitrate, and the halides, and as organomercurials generated by microbial and anthropogenic processes [1]. Mercury also has a zero valent form (Hg0), which is stable at standard temperatures and pressures as a dense liquid with a highly volatile monoatomic vapor [1]. Approximately 75 % of mercury released to the environment comes from anthropogenic sources, primarily emissions from fossil fuels, and urban and industrial waste disposal sites, although natural sources such as volcanoes also contribute mercury to the environment [2]. Historically, mercury has been used in classical and medieval medicine and alchemy [3]. Modern uses include batteries, switches, thermostats, electrodes (e.g. in the chlor-alkali process) and medical devices, including the Hg-Ag amalgam of dental restorations, a major source of chronic exposure in humans [1, 4, 5]. Two other very direct sources of human exposure are through the use of mercury in artisanal gold mining [6] and consumption of fish containing methyl-mercury [4]. Chronic and acute mercury exposure in humans can result in neurotoxicity, nephrotoxicity, immunotoxicity, hepatotoxicity, gastrointestinal toxicity, and neonatal development problems [4, 7–9]. The organomercurials, methylmercury and ethylmercury (a component of the common disinfectant, merthiolate, also known as thimerosal), are considered primarily neurotoxic and both inorganic forms, Hg0 and Hg2+, are considered primarily nephrotoxic [10]. The biochemical basis for these differences is unclear.
As with other ubiquitous environmental electrophiles such as arsenic and lead, there is no single biochemical target of Hg damage. Mercury has a strong affinity for sulfur ligands [11, 12], so the expected cellular targets for mercury are low molecular weight thiols (RSH) involved in intracellular redox homeostasis [13] and the thiol groups of proteins [10]. The major low molecular weight thiol in animals and many bacteria is glutathione (GSH), which acts as an intracellular redox buffer [14]. Approximately 92 % percent of human proteins have one or more cysteines [15]. In vitro, mercury damages enzymes with an active site cysteine [16], displaces metal ion cofactors [17], disrupts structural stability [18] and forms a stable cross-link between intra- and inter-protein cysteine residues [16, 19]. Mercury reacts even more strongly with the selenol (RSeH) of the rare amino acid selenocysteine, found across all domains of life [20, 21].
Cells maintain optimum available concentrations of essential metals homeostatically [22, 23], and disruption of these balances by a toxic metal can damage many processes [24]. Approximately half of all enzymes in the six Enzyme Commission (EC) functional classes use metal cofactors [25–27]. Transition metal cofactors are often coordinated by protein cysteine residues and are vulnerable to displacement by mercury with consequent loss of protein function. Despite many studies on individual enzymes and cellular processes, there has been no comprehensive study of the bulk effects of Hg on the thiol pool and metal homeostasis in any organism. As part of a larger proteomics project to define the mercury exposome [28], we examined the effects of brief, acute exposure of growing cells to inorganic and organomercurials (Fig. S1) on the cellular content of thiols, of essential metals, and of free iron and iron-binding proteins in E. coli K-12 MG1655.
Materials and methods
Preparation of cells
For each experiment E. coli K-12 MG1655 was subcultured from cryostorage on Luria–Bertani (LB) agar and grown overnight at 37 °C. A half-dozen well-isolated colonies were used to inoculate a 50 ml starter culture of Neidhardt MOPS minimal medium [29] supplemented with 20 mg/L uracil and 5 mg/L thiamine, which was then incubated at 37 °C with shaking at 250 rpm for 18 h. Neidhardt MOPS minimal medium was selected as it is the standard medium for most ‘omics work in E. coli using K-12 MG1655 strain. The saturated starter culture was diluted 1:40 to initiate the experimental culture, which was then incubated at 37 °C with shaking at 250 rpm. At OD595 of approximately 0.6, the culture was split into four 450 ml aliquots in Fernbach flasks. For each experiment a defined concentration of each mercurial (i.e. a compound containing mercury) or oxidant was added to all but one culture aliquot (the unexposed control) and all were incubated for 15 min or 30 min at 37 °C, with shaking at 250 rpm. All data presented are from 30-min exposures except for proteomics data (Tables 2 and S3–S8) and for BODIPY-protein-thiols data in Fig. 2, which are from 15-min exposure cultures. Cultures were harvested by centrifugation at 17,700×g at 4 °C. Each cell pellet was suspended in ice-cold MOPS minimal medium at one 100th the original culture volume (100×), and then dispensed as 1 ml aliquots. The cells were washed by centrifuging at 10,600×g for 4 min at 4 °C, suspending in 1 ml of ice-cold 20 mM Tris–HCl (pH 7.4), repeating centrifugation, and finally suspended in 500 µl (200×) of 20 mM Tris–HCl (pH 7.4) for cryostorage at −70 °C until they were used in specific assays as described below. Where assays required lysates rather than intact cells, the frozen cell suspensions were thawed on ice, diluted to 50× with 20 mM Tris–HCl (pH 7.4) and lysed 3 times by French Press (10 K psi, 4 °C).
Quantifying protein
The protein concentrations of the crude French Press lysates were quantified by the Bradford method [30] using Coomassie Plus™ reagent (Pierce). The number of cells was estimated based on protein constituting 16 % of the cell wet weight and the wet weight of one E. coli cell being 9.5 × 10−13 g [31]. Where intracellular concentration is reported, a cell volume of 6.7 × 10−16 L/cell was used for calculations [31].
Quantifying total cellular thiols
Total thiols in French Press cell lysates of cultures exposed to mercurials (or not) for 30 min, were determined by reaction with 5,5′-dithiobis(2-nitro-benzoic acid) (DTNB; Sigma) using the extinction coefficient at 412 nm for thionitrobenzoate (TNB) of 13,600 M−1 cm−1 [32]. The total thiol to Hg ratio was determined based on the calculated molecules of thiol per cell in the unexposed control, relative to the total atoms of mercury quantified by ICP-MS per cell in exposed cultures. In order to measure thiols of proteins in the native state available for interaction with the mercurials, urea was not used here.
Detecting protein thiols
Protein thiols in fresh French Press lysates of mercurial-exposed cultures were treated (or not) with 10 M urea, and covalently labeled in the dark, at room temperature, by reaction for 60 min with BODIPY® iodoacetamide (BODIPY-I, BODIPY® FL C1-IA, N-(4,4-Difluoro-5,7-Dimethyl-4-Bora-3a,4a-Diaza-s-Indacene-3-yl)Methyl)Iodoacetamide) (Invitrogen) using a twofold molar excess over total lysate thiol concentration measured in the unexposed control culture (see Table 1 footnote). Unreacted BODIPY-I was removed with a Sepharose gel (BioGel P-2, BioRad) spin column and eluted protein was quantified by the Bradford method [30]. The excitation spectrum of BODIPY does not overlap with the Bradford assay and controls were performed to confirm this (data not shown). A constant protein mass of BODIPY-tagged cell lysate was separated by SDS-PAGE on a 12.5 % Next-Gel® (Amresco) and, after rinsing with deionized water, the protein-associated BODIPY fluorescence was recorded with a GE Typhoon Trio (488 nm excitation, 520 nm bandpass 40 emission filter, 375 V photomultiplier tube (PMT), 200 µm resolution, normal sensitivity). Lastly, to measure total protein intensity in each gel lane, the gel was stained with Imperial™ protein stain (Pierce), destained in deionized water overnight, and imaged with Typhoon (Ex: 633 nm excitation, Em: 670 nm bandpass BP30 emission filter, PMT: 750 V, 200 µm resolution, normal sensitivity), which also served as confirmation that an equal mass of protein was loaded in each lane. Using GeneTools (Syngene, Inc), the summed intensity (fluorescence or Coomassie stain) for each lane of a mercurial-exposed lysate was compared to that of the unexposed lysate to estimate the loss of BODIPY reactivity or of bulk protein in each exposure condition. Densitometric protein profiles from exposed and unexposed cultures were also overlaid to look for gain or loss of individual fluorescent or Coomassie-stained bands.
Quantifying free iron by electron paramagnetic resonance (EPR)
A 1 ml aliquot of 100× intact cells, from 30-min exposure cultures, was treated with 100 µl of 100 mM of cell-impermeant diethylene triamine pentaacetic acid (DTPA; Sigma) to block iron uptake from the medium and with 100 µl of 200 mM cell-permeant deferoxamine mesylate salt (DF; Sigma). The iron chelator deferoxamine does not disrupt protein-bound iron and will only bind free (i.e. uncomplexed iron that is not bound by proteins) intracellular Fe(III) and Fe(II) that is oxidized in the presence of the iron chelator and molecular oxygen to form the S = 5/2 Fe(III):DF complex, which exhibits an intense EPR signal at g = 4.3 [33]. An identical aliquot was not treated with DTPA/DF and both were incubated at 37 °C, 250 rpm for 15 min and then centrifuged at 10,600×g for 4 min at 4 °C. The cell pellets were washed with 1 ml of ice-cold 20 mM Tris–HCl (pH 7.4) and each pellet was suspended in 300 µl of 20 mM Tris–HCl (pH 7.4) in 10 % (vol/vol) glycerol and cryostored at -70 °C until EPR analysis. For EPR analysis cell suspensions were thawed on ice and loaded into quartz EPR tubes and re-frozen in liquid nitrogen. EPR standards of the Fe(III):DF complex were generated by reacting 100 µM FeCl3 in 20 mM Tris–HCl (pH 7.4) and 20 mM DF for 15 min at 37 °C, 250 rpm. The concentration of the Fe(III):DF chromophore was quantified at 420 nm using an extinction coefficient of 2865 M−1 cm−1 [33].
Dilutions of the Fe(III):DF complex were made in 20 mM Tris–HCl (pH 7.4) in 10 % glycerol for 0, 10, 20, 30, 40 and 50 µM standards. X-band (~9.6 GHz) EPR spectra were recorded at −203 °C (70 K) using an ESP-300D spectrometer (Bruker, Billerica, MA), equipped with an ESR 900 helium flow cryostat (Oxford Instruments, Concord, MA). The Fe(III):DF concentration of each sample was determined from the standard curve under non-saturating conditions by using peak-to-trough height of the isotropic EPR signal at g = 4.3.
Observing mercury adducts of E. coli iron-binding proteins by LC–MS/MS
Cultures were prepared as above for 15 min exposure to 40 µM PMA, 160 µM merthiolate or 20 µM mercuric acetate, but harvested cells were suspended in ammonium bicarbonate buffer amended with iodoacetamide (IAM) to prevent redistribution of mercury adducts by exchange with free thiols during preparation for liquid chromatography-coupled mass spectrometry (LC–MS/MS) proteomic analysis [28] (Zink et al., manuscript in preparation). Gene Ontology (GO) terms in UniProtKB [34] were used to generate a list of all encoded E. coli proteins that bind iron alone, in iron-sulfur clusters, or in heme groups (Table S1 and S2). This list (current as of June 2015) was searched against the results of LC–MS/MS proteomes obtained from cultures exposed to PMA (3 biological replicate proteomes, Table S3), merthiolate (MT) (1 proteome, Table S4) or mercuric acetate (1 proteome, Table S5) (full datasets to be published in Zink et al., manuscript in preparation) to identify iron-binding proteins whose cysteines formed stable adducts with these compounds. All proteome summaries contain data from corresponding no-mercury control samples, which are included in total spectral counts.
LC–MS/MS peptide hit lists were generated by SEQUEST using the E. coli genome sequence (GenBank: U00096.2) and allowing alkylation by IAM or adducts of Hg, phenylHg (PhHg) or ethylHg (EtHg) as modifications of cysteine. SEQUEST results were re-scored by MS-GF [35], and SEQUEST matches to Hg and PhHg and EtHg adducts were additionally re-scored by PeptideProphet [36] to maximize true identifications of Hg adducts. Filtering criteria were selected based on false discovery rates (FDR) computed from searches against a reversed sequence database as decoys. The primary filter for spectral matches was MS-GF’s spectral probability, which we required to score less than 1.6E−10 for acceptance of any peptide match. This threshold yielded a FDR of 0.001 over all spectra. To eliminate false discoveries that are singletons, we further improved accuracy by both requiring more than one spectrum for any peptide and, for proteins with only a single observed peptide, requiring a MS-GF spectrum probability less than 1E−11 and at least one tryptic end (semi or fully tryptic). For matches to peptides modified by Hg, we relaxed the MS-GF spectrum probability threshold to 5E−7, but required PeptideProphet’s probability to be greater than 0.8 for acceptance. Both thresholds yielded a high FDR when used alone, but when combined and with two additional criteria that peptides be fully tryptic and from proteins passing by the MS-GF spectral probability filter, these criteria produced zero matches to Hg-modified decoy sequences (FDR = 0.0). As an alternative estimate of FDR focused on incorrect modifications rather than on incorrect sequences, we used “Hg adducts” detected in the Hg-free samples as a distinct estimate of FDR. This yielded an FDR of 0.009 for spectra identified as Hg-modified. Detailed qualitative (Zink et al., in preparation) and quantitative (Polacco et al. in preparation) analyses of mercurial modifications of the complete E. coli proteomes are underway and will be submitted elsewhere.
Quantifying bulk cellular inorganic elements
One ml cryoarchived aliquots of 100× concentrated intact cells from each exposure condition were thawed on ice and brought to 5 ml total volume with HPLC grade water. They were then diluted with concentrated trace metal grade nitric acid including a 1 ppm gold background and digested by microwave in sealed Teflon containers. Metal concentrations in the digested solutions were determined with a Perkin-Elmer ELAN 9000 ICP-MS using internal standards and intermittent blanks to exceed requirements for EPA Method 3051A (http://www.epa.gov/osw/hazard/testmethods/sw846/pdfs/3051a.pdf) and 6020B (http://www.epa.gov/wastes/hazard/testmethods/sw846/pdfs/6020b.pdf). These analyses were carried out at the University of Georgia Laboratory for Environmental Analysis (http://www.uga.edu/lea).
Identifying bulk cellular mercury ligands
Mercury was added to a known amount of growing cells and incubation was continued for 30 min followed by harvest, concentration and cryostorage as above. A 500 µl aliquot of 200× concentrated cells for each exposure condition was thawed on ice and 8 µl was loaded into 5 wells of an XAS microcuvette for each exposure condition examined and flash frozen in liquid nitrogen. Pure standards of mercury compounds mixed with glutathione (or not) were prepared at room temperature and 8 µl was loaded into each of 5 wells of an XAS cuvette and flash frozen in liquid nitrogen.
Mercury L3-edge data were collected at −263 °C (10 K) beamlines 9–3 and 7–3 at the Stanford Synchrotron Radiation Laboratory (SSRL), with the SPEAR storage ring operating in a dedicated mode at 3.0 GeV and 50 to 100 mA. An Si[220] double crystal-monochromator and a 30-element Ge solid state X-ray fluorescence detector were employed for data collection. No photoreduction was observed when comparing the first and last spectra collected for a given sample. The first inflection of a Hg-Sn amalgam standard was used for energy calibration. Extended X-ray absorption fine structure (EXAFS) analysis was performed with EXAFSPAK software (http://www-ssrl.slac.standford.edu/exafspak.html) according to standard procedures [37, 38]. Fourier transforms (FT) were calculated with sulfur-based phase-shift correction. Theoretical and phase-shift functions employed in simulations were generated with FEFF 8.2 code [39, 40]. Curve fitting analysis was performed as described previously [41].
Results
HgCl2 blocked total and protein-associated thiols more effectively than did phenylmercury or merthiolate
Blockage of cellular thiols after 30 min exposure
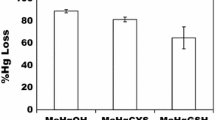
As measured by Ellman’s reagent (DTNB), lysed cells from unexposed cultures had an average bulk thiol content of 3.59E+06 molecules/cell ±1.54E+05 (Table 1). The two organomercurials examined, PMA and MT, were used as surrogates to methylmercury, but are also of public health interest in their own right. The organomercurial PMA at 40 µM in the media, accumulated to levels that blocked all DTNB-detectable thiols. Merthiolate at 160 µM in the media, only blocked approximately 89 % of total thiols. In contrast, exposure to as little as 8 µM HgCl2 in the media resulted in a 46 % loss of reactive thiols and at 20 µM HgCl2 or above, cellular thiols were undetectable. Thus, neutral, bivalent, HgCl2 is more effective per mole in blocking thiols in intact growing cells than larger lipophilic, monovalent PMA or MT. Note also that PMA and Hg(II) accumulated in cells to levels in excess of the measured thiol content of the cells (rightmost column in Table 1); this is examined further in the last section of the “Results”.
The fluorescent thiol-reactive dye, BODIPY-iodoacetamide was used to generate a qualitative SDS-PAGE “fingerprint” of total cell lysate proteins with reactive thiols. The profiles of BODIPY-tagged proteins, not treated with urea, were nearly indistinguishable from each other (Fig. 1). Thus, during 30 min exposure each compound was able to enter intact cells and form stable adducts to protein thiols preventing subsequent reaction with the fluorescent probe BODIPY.
Differential effects of Hg(II), PMA, or merthiolate (MT) on total detectable protein thiols. Lysates of cells exposed (or not) for 30 min during growth to mercurials as indicated were reacted with BODIPY-I. Their proteins were separated by SDS-PAGE and imaged by fluorescence (GE Typhoon: Ex: 633 nm, Em: 670BP30, PMT: 750 V, 200 µm resolution, normal sensitivity). Lanes presented are from a single gel that was trimmed for this figure; lanes not shown were identical to these, confirming that the additional 15 min at 37° for DF and DTPA treatment had no effect on BODIPY detection of protein thiols. Fluorescence Intensities (FI) for each condition are normalized to the corresponding unexposed cells (lane 1). See “Materials and methods”
Blockage of protein-bound thiols after 15 min exposure
Shorter exposure time revealed differences between the mercurial compounds in BODIPY-iodoacetamide tagged lysate profiles of cells exposed to mercurials for only 15-min (Fig. 2). Urea treatment of the lysate prior to reacting with BODIPY-I uncovered additional thiols, presumably in the interior of proteins (even numbered lanes). A range of concentrations for each mercury compound was tested to determine exposure conditions that would result in intracellular accumulation high enough to bind protein thiols and allow for detection of mercury adducts in proteomics analysis. Note that BODIPY-iodoacetamide was chosen over its maleimide derivative for its preference for cysteine thiols [42]. Faint gel bands observed in lysates lacking any detectable thiols by Ellman’s assay (Table 1) likely arose from BODIPY’s acetamide’s weak reactivity with the more abundant amino acids histidine [43], lysine [44], and tyrosine [45].
Effects of Hg(II), PMA, or merthiolate (MT) on total detectable protein thiols. Lysates of cells exposed (or not) to mercurials as indicated for 15 min during growth were reacted (or not) with 10 M urea and then with BODIPY-I. Their proteins were separated by SDS-PAGE and imaged by fluorescence (GE Typhoon: Ex: 633 nm, Em: 670BP30, PMT: 750 V, 200 µm resolution, normal sensitivity). Images from several gels prepared under the same conditions were compiled for this figure. Total densitometric lane intensity of unexposed cultures varied by 17 % s.d. without urea (e.g. lane 1) and by 13 % s.d. with urea (e.g. lane 2). Total densitometric lane intensity for each condition was normalized to the corresponding lane intensity of unexposed cells. see “Materials and methods”
The total fluorescence intensity in each lane was compared densitometrically to the corresponding urea-treated or untreated lane for the lysate from the unexposed condition. No unique BODIPY-reactive protein bands appeared or disappeared upon mercury exposure (data not shown). Rather, BODIPY-reactive proteins decreased uniformly with increasing mercury exposure. However, there was no change in intensity of bulk protein band patterns as detected by Coomassie Blue (data not shown) indicating the mercurial-provoked loss of BODIPY-reactivity is not caused by non-specific loss of proteins through precipitation or aggregation.
In lysates of cells exposed to 20 µM (lane 3) or 80 µM HgCl2 (lane 5), BODIPY-protein fluorescence decreased by 38 and 39 %, respectively, compared to the corresponding unexposed cell lysate (lane 1). Urea treatment of the unexposed lysate (lane 2) increased BODIPY fluorescence (relative to lane 1) by unmasking buried cysteine thiols. In contrast, urea treatment of lysates of Hg-exposed cells yielded very low BODIPY fluorescence (lanes 4 and 6 compared to lanes 3 and 5, respectively), indicating that in the intact cells these nominally buried cysteines had either been blocked directly by Hg(II) or possibly oxidized to disulfides by reactive oxygen species provoked by Hg(II) inhibition of respiratory proteins (see Fe-release and Fe-binding proteome results below).
Exposure to the larger aromatic, monovalent mercurial, phenylmercury acetate (PMA) blocked BODIPY access to only 24 % (20 µM, lane 7) and 29 % (40 µM, lane 9) of protein thiols in lysates without urea compared to lysates of cells not exposed to mercurials or urea (lane 1). Urea treatment of lysed PMA-exposed cells also showed more modest blockage (greater BODIPY fluorescence) at 20 µM PMA (lane 8) than the corresponding urea-treated lysate of cells exposed to 20 µM HgCl2 (lane 4). However, the urea-treated cells exposed to 40 µM PMA (lane 10) did suffer a decrease in BODIPY fluorescence equal to the 20 µM HgCl2-exposed cells, consistent with their respective valences. This suggests that neutral HgCl2 enters the cell no less rapidly than hydrophobic phenylmercury, which may also be present as the chloride in this minimal medium.
Exposures to the large, negatively charged merthiolate (MT; ethyl-mercury thiosalicylate), did not block protein cysteines from reaction with BODIPY at 40 µM (lane 11) or 80 µM (lane 13) and blocked only weakly at 160 µM (lane 15) (compared to lysates from cells exposed neither to mercurials nor urea, lane 1). Urea treatment showed that just 11, 14 and 19 % (lanes 12, 14, and 16 compared to lane 2) of BODIPY-reactive thiols had been blocked by merthiolate in intact cells. This weak blockade of protein and cellular (Table 1) thiols by MT is consistent with its uptake impediments (size and negative charge) and the strength of the S–Hg bond with its thiosalicylate ligand (Kform of 1038, [46]). Thiosalicylate is thermodynamically less likely to be displaced from ethylmercury by protein or cellular thiols than the more weakly associated acetate of PMA (Kform 108, [47]) or chloride (Kform 1014, [48]) of Hg(II).
Thus, in this bulk qualitative “fingerprint” analysis with BODIPY, these three Hg compounds differed in uptake rate, but did not differ significantly in their ability to react with cellular thiol targets that they ultimately blocked, though whether the blockage was directly by mercurial ligands or indirectly by oxidation of the thiols upon mercurial-provoked generation of reactive oxygen species (ROS) is not distinguishable by these methods.
Mercurials released protein-bound Fe(II)
Iron is a redox active metal, so the concentration of intracellular uncomplexed iron, i.e. iron that is not bound to proteins, is normally kept low at 15–30 µM by sequestration via storage proteins [33]. The labile iron pool [i.e. protein-bound but readily releasable Fe(II)] in E. coli is approximately 100 µM and is largely comprised of iron-sulfur clusters in proteins of several essential cellular pathways [49]. Heme–iron and Fe(III) in storage proteins, such as bacterioferritin, are not considered labile under aerobic conditions [50]. In agreement with published values [33], using EPR to observe Desferal-complexed Fe(III) we found uncomplexed iron concentrations of 20–25 µM in intact cells not exposed to mercurials (Fig. 3). Cells exposed to the common oxidants, 4 mM H2O2 or 8 mM tert-butyl hydroperoxide, had 15–25 % more free iron [non-protein-bound Fe(III)] than unexposed cells. Both organomercurials were more effective on a molar basis at releasing iron than were the common oxidants. MT at 25-fold lower concentration released a similar amount of iron as H2O2 and PMA at 100-fold lower concentration than H2O2 nearly doubled the free iron content. Inorganic mercury, HgCl2, increased free iron more dramatically on a molar basis than both the common oxidants and the organomercurials. As little as 16 µM HgCl2 caused a 120 % increase in free iron and 80 µM HgCl2 caused an average 220 % increase in uncomplexed iron, freeing as much as 80 % of the estimated labile iron pool. The increase in free iron in the cell could have generated reactive oxygen species via Fenton chemistry, but mercury interference with the OxyBlot assay precluded determination of oxidative damage to proteins via carbonylation (data not shown).
Increases in free intracellular iron levels in cells exposed to mercurials and common organic oxidants. The increase in intracellular free iron is represented as the average X-fold increase in the Fe(III):DF complex EPR signal at g = 4.3 for each stress condition relative to the unexposed control whose average free iron concentration was 24.4 µM (±4.9 µM). Error bars are standard deviation of biological replicates. Replicates for each condition were: Unexposed (6): H2O2 (2); t-BuOOH (2); MT (3); PMA (3); 16 µM HgCl2 (3); and 80 µM HgCl2 (5)
Mercurials formed stable adducts with cysteines of iron-sulfur centers
Using shotgun LC–MS/MS proteomics modified to detect protein–mercury adducts in mercurial-exposed cultures, we observed 78 of E. coli’s 214 encoded Fe-binding proteins based on gene ontology in UniProtKB database (Table S2) by one or more peptides each (50,870 total spectra) (Table S6). We detected cysteine-containing peptides from 46 iron-binding proteins of the 75 that encode at least one cysteine in the 4571 spectra (9 % of the total spectra from Fe-proteins). Thirty-four cysteine sites from 20 different Fe-binding proteins (416 spectra total, 9 %) were observed with mercury adducts (Tables S6 and S7). Some Hg adducts were observed in PMA-exposed cultures which likely arose from spontaneous dephenylation of the PhHg adduct in peptides with multiple cysteines, possibly via a mechanism analogous to MerB [51, 52].
We operationally defined a cysteine residue that was modifiable by IAM (yielding the carbamidomethyl- or CAM-adduct) in the no-mercury control condition as one that would also be available for modification in mercury-exposure conditions. Indeed, cysteine sites yielding many CAM-adducts in the no-mercury condition were generally seen with PhHg- or Hg-adducts when exposed to mercurials. At Cys positions showing <10 CAM adduct spectra mercurial adducts were rarely observed. This could arise from unknown intrinsic differences in the detectability of a given peptide by MS when it bears chemically distinct adducts. Thus, this more fine grained, but still qualitative, analysis was consistent with the above bulk analyses with DTNB and BODIPY-I (Table 1; Figs. 1, 2), i.e., cysteines modifiable by IAM, were also available for modification by mercurials (Table S6).
Eleven of the 20 Fe-binding proteins observed with RHg- and/or Hg-adducts had a mercury bound to a cysteine site that is directly involved in coordinating non-heme iron or iron present in an Fe-S cluster (Tables 2 and S7). Nine of these sites are in Fe-S centers or are involved in cluster assembly, two proteins use an iron ion as a cofactor and one protein has not been structurally characterized. Thus, mercurials can displace Fe from many different proteins as also reported for a specific example, the 4Fe-4S dehydratases [53]. Our LC–MS/MS identification of Hg-adducts reveals for the first time that in vivo mercury compounds make stable adducts with the vacated Fe-S center cysteines, potentially impeding the repair of those Fe-S centers. In addition, the chemistry dictates that once mercury is bound to thiols (log β 2 = 40–45 of HgL2 thiol complex [11, 12, 54]), this type of modification is not easily repaired and iron (log β 2 = 18.8 of Fe-(cys)2 [55]) cannot simply displace the mercury from the thiol. Observable adducts of inorganic Hg(II) appear to be biased toward two-cysteine peptides that offer the stability of chelation. Three such proteins are conserved in higher organisms, IscU and MsrB in mitochondria specifically (see footnote in Table 2).
Proteomics data were also searched for other transition metal binding proteins to determine if mercury can target these proteins (Table S8). There were few- to no-mercury adducts to proteins that bind Co (1), Cu (1), Mn (5) or Ni (0) compared to Zn (27 distinct proteins). This is consistent with Zn being a group 12, soft, divalent metal with thiophilic coordination preferences similar to Hg, whereas the others use N, O, or thioether (i.e. methionine) sulfur ligands as often or more often than thiol sulfur (i.e. cysteine) that Hg prefers. ROS-induced protein modifications were not searched in the proteomics datasets, due to difficulty in distinguishing true in vivo induced modifications versus artifacts arising from sample processing without special considerations [56], which was not a research goal of this proteomics project.
Mercurials disturbed the electrolyte balance
Of nine essential metals examined by ICP-MS in intact cells (Fig. 4), only the bulk alkali metal electrolytes, Na+ and K+, changed significantly after exposure to mercurials. In cultures exposed to 80 µM HgCl2 the K+ content decreased 62 % and the Na+ content increased 200 % relative to unexposed cells. Similar changes were observed in cultures exposed to 40 µM PMA, 16 µM HgCl2 and 160 µM MT (Fig. 4) but with lower magnitude. These K+ and Na+ changes are consistent with mercurial disruption of thiol homeostasis and consequent K+ efflux by the GSH-responsive K+/H+ antiporter KefC [57] resulting in lower intracellular pH [58]. The increase in Na+ could be a consequence of Na+ uptake by the NhaA (Na+/H+ antiporter) in response to the low intracellular pH provoked by KefC importing H+ as it ejects K+ [59–61].
Effect of mercurials on intracellular levels of essential metal ions. Exponential phase MG1655 cells were unexposed (black) or exposed to 40 µM PMA (dark grey), 160 µM MT (medium grey), 16 µM HgCl2 (light grey) or 80 µM HgCl2 (off-white) for 30 min, harvested, washed, and assayed for nine essential elements by ICP-MS. Inset K+ and Na+ content on smaller scale. Error bars represent standard deviation of biological replicates: Unexposed (7); PMA (4); MT (4); 16 µM HgCl2 (3); 80 µM HgCl2 (6). Significance (*) was determined by t-test comparison of the means at the 95 % confidence level with two-tailed p-values equal to <0.05
Interestingly, although EPR (Fig. 3) showed a large increase in uncomplexed iron on exposure to 16 or 80 µM HgCl2, total cellular iron content did not change significantly indicating little or none of the free Fe was lost from the cells. Thus, the putative defensive ferrous ion efflux activity of cation diffusion facilitator FieF [62] may be non-functional under acute mercurial exposure. Other Fe homeostasis functions including down-regulation of iron uptake [63] and induction of Fe(III) storage proteins [62] may also be non-functional. Transporters for the other metal cations are energy-dependent [64], so their efflux may have been prevented by immediate inhibition of these processes by the mercurials. No other metals experienced statistically significant differences in intracellular concentration from the unexposed under experimental conditions used.
Cells bound mercury compounds in large excess over their total available thiol ligands
Bulk cellular binding of mercury compounds after 30 min exposure
The total thiol content of washed cells not exposed to mercurials averaged 3.59E+06 molecules/cell (approximately 9–10 mM, assuming a cytosolic volume of 6.7 × 10−16 L/cell [31] ) (Table 1). Experimental conditions used to measure cell-bound metals normally assume both uptake and efflux and therefore derive equilibrium values. However, the nearly covalent nature of Hg–S bonds, especially in bis-coordinated complexes, renders them essentially irreversible unlike less thiophilic transition metals. Likely owing to slow uptake even at 160 µM in the medium the large, monovalent anionic MT bound only 70 % of available thiols. However, both PMA and inorganic Hg were stably bound in considerable excess over the total cellular thiol content at much lower exposure. Cells exposed to just 40 µM of the smaller aromatic, monovalent PMA bound almost 1.5-fold more of it than the measured thiol groups available. Even cells exposed to 16 µM inorganic HgCl2 bound a 1.23:1 molar excess of it over measureable thiols. Since Hg is bivalent, these cells accumulated 2.46-fold more Hg than needed to occupy all of their available thiols. At 80 µM HgCl2 the cells bound a 3.4:1 molar excess of Hg to measured thiols or 6.8-fold more Hg than needed to saturate all cellular thiols. Thus, even when normal metal uptake systems are likely shut down by the toxic metal, a variety of cellular ligands competed well with weaker ligands in the medium for the two neutral, membrane-permeant compounds, HgCl2 and phenylmercury (probably as the chloride given the medium composition). Intrigued by these findings, we used X-ray absorption fine structure (EXAFS) spectroscopy to learn what additional ligands mercury was taking in E. coli cells.
Excess cell-bound mercury takes nitrogen and/or oxygen ligands in addition to sulfur
EXAFS reports the type, number, and distance of elements coordinated to a specific metallic element [37]. We exposed actively growing cells for 30 min to a range of HgCl2 concentrations from under-saturation to over-saturation of cellular thiols to discern novel ligands arising as Hg-binding sites exceeded the cellular thiol pool (Fig. 5 and Table S9 for fit details). Cells exposed to 10 or 20 µM HgCl2 fit best to a model of two sulfur ligands at 2.34 Å radius as expected based on Hg/thiol ratios observed in Table 1. At higher HgCl2 concentrations that exceeded the cellular thiol pool, EXAFS heterogeneity increased indicating Hg coordination to other ligands near 2.0 Å consistent with nitrogen, oxygen, or carbon (low mass elements not distinguishable by EXAFS). Nonetheless, Hg:S coordination remained dominant and was distinguishable from Hg:Cl (Fig. 5a) and from mercurial complexes with model sulfur (glutathione, Fig. S3b and S3d and S4), oxygen (acetate, Fig. S4), or nitrogen (imidazole, Fig. S5) compounds by near edge and FT features. At 40 or 80 µM HgCl2 exposure the shorter and broader Fourier transform peaks yielded mixed, non-integral fits (Fig. 5d). Although oxygen and carbon gave similar first shell fits to those for nitrogen (data not shown), nitrogen is softer than oxygen and is the more likely biological ligand given mercury’s known preferences for bis-coordination with the imino nitrogens of nucleotide bases [65, 66] and histidine [67]. A Hg:C signal could have arisen from oxymercuration of unsaturated lipids but this remote possibility was ruled out by ICP-MS quantification of Hg in cellular fractions prepared by a standard lipid extraction [68] (data not shown).
Changes in cellular ligands as Hg concentration is increased. HgCl2 (10 mM, grey) was added to a known amount of growing cells at 10 µM (blue), 20 µM (green), 40 µM (red), or 80 µM (pink). Panels are the near edge (a) and EXAFS (b) spectra of the concentrated washed cell suspensions and the HgCl2 standard. Panel c is the corresponding Fourier transforms of the EXAFS data in b; panel d contains the EXAFS fitting results. Best Fit subscripts denote the number of scatterers per metal atom. R as is the observed metal-scatterer distance. σ 2as is the Debye–Waller or temperature factor
Cells exposed to 40 µM PMA best fit a one carbon (2.05 Å) and one sulfur (2.32 Å) structure reflecting the carbon of the phenyl group and a sulfur of a cellular thiol group (Fig. S6). Although cells exposed to 40 or 80 µM PMA bound an almost 1.5-fold molar excess of PhHg (Table 1), its native oxygen from acetate and carbon ligand to phenyl group precluded resolution of cellular C, O, or N ligands by EXAFS. However, cellular thiol ligands were distinguishable by EXAFS when they replaced the phenylmercury’s acetate oxygen ligand (Fig. S6c). In contrast, cells exposed to 160 µM MT fit only to one carbon (2.10 Å) and one sulfur (2.36 Å) each, reflecting the ethyl group carbon and a sulfur from either MT’s thiosalicylate moiety or the cellular thiol pool (Fig. S7). Note that HgCl2 and PMA reacted with the model compound imidazole to form a Hg:N bond (Fig. S5) but no Hg:N bond was observed when merthiolate was mixed with imidazole, indicating that the thiol of merthiolate was not displaced by imidazole. The N3 imino nitrogen of imidazole is not sufficiently nucleophilic to displace the strong S ligand of thiosalicylate and the covalent C:Hg bond of ethylmercury is not exchangeable.
Discussion
Knowing how mercury compounds differ in their basic biochemical interactions in vivo is fundamental to understanding the damage they do to cells, how cells recover—or not—from that damage, and what interventions can prevent or minimize the damage and accelerate recovery. This study is the first to compare the effects of inorganic and organic mercury compounds at the biochemical, physiological, and proteomic levels in any model organism.
Relevance to environmental exposures
The work described here is part of a larger proteomics study carried out in a multi-user, production-scale, high-throughput proteomics facility and for safety reasons we used two common antiseptics, PMA and merthiolate (MT), as surrogates for the often mentioned neurotoxin, methylmercury. PMA and MT are also of public health interest in their own right. Inorganic mercury concentrations employed here were within the range that bacteria would experience in the GI tract after installation or removal of dental amalgam fillings [69] and in highly contaminated environments, such as mercury mines [6]. Natural bacterial exposures to the organomercurials are more difficult to estimate. Methylmercury was long used as a seed grain fungicide, and phenylmercury and merthiolate were previously used in contact lens cleaning solutions [70, 71] and vaginal douches [72] and the latter was also used for minor skin wounds and as a surgical scrub [73], but there are no data on the resulting ambient concentrations. The use of thimerosal (the same as MT) in vaccines was questioned in 2001 [74] and it has gradually been removed from pediatric vaccines administered in the United States, but is still included in pediatric vaccines administered in developing countries and in adult vaccines worldwide [75].
Effects of mercury compounds on bulk properties of the cell
Blocking total and bulk protein thiols
In brief exposures the relative effectiveness of each compound for blocking cellular thiols on a per mole basis was HgCl2 > PMA > MT (Table 1; Figs. 1, 2). Monovalent PMA was roughly half as effective short term as bivalent Hg(II) in blocking bulk cellular or protein thiols, suggesting that on short exposure the relative efficiency of thiol blockage simply reflects the valence of the mercurial. Uptake and accumulation of mercury were aided by the fact that cells were grown in a defined minimal medium. The shaking required for aerobic growth ensured distribution of mercury throughout the culture and the growth medium contains no strong competitive ligands for mercury compounds apart from chloride, so the cells become a thermodynamic sink for mercury due to their abundance of suitable intracellular ligands. Neutral HgCl2 [76] and PMA, which is likely phenylmercuric chloride due to the high concentration of chloride in the medium (~55 mM), were likely taken up with similar efficiency by passive diffusion [77]. In contrast, despite its generally hydrophobic character the predominantly negatively charged carboxylate group of merthiolate (pK a ~4.2) under the conditions of these studies (pH 7.4) (Fig. S1) is expected to slow its entry into the cell. In longer term exposure (Fig. 1) thiol blockage by all three compounds is nearly identical, although some of this may result from mercurial-provoked ROS.
E. coli exports glutathione which reaches micromolar concentrations in the periplasm [78] where it may function in disulfide bond formation [79]. However, E. coli does not readily take up mercury bound to glutathione [78, 80]. Although cysteine may increase bioavailability of inorganic mercury, it has little effect on organomercurial uptake [77, 80], likely because in aerobic culture extracellular thiols will be disulfides. Our finding that cells accumulate inorganic mercury and phenylmercury in excess of measured available thiols suggests passive diffusion of neutral species driven first by intracellular thiols and then by intracellular non-thiol ligands. This is more probable than active uptake of mercury in MG1655 which does not carry a typically plasmid-borne mercury resistance (mer) locus [1]. In addition, this quantitative measurement of accumulation reveals that inorganic mercury continues to accumulate as the concentration increases, while PMA appears to plateau at less than twofold excess and much higher concentrations of merthiolate do not saturate the cellular thiol pool within the same exposure time.
Once mercurials enter the cytoplasm 5–10 mM glutathione will readily displace (logKform = 35–40) the weakly associated chloride or acetate counterions and even thiosalicylate, which has a less basic, lower affinity thiolate [81, 82]. The resulting derivatives of Hg(II), phenylmercury, and ethylmercury can readily swap their glutathione ligands [11] for protein cysteine thiols. Organisms have evolved pathways to repair damage to oxidized protein thiols [83], but mercury bound to a protein cysteine can only be rapidly and effectively removed by the proteins of the mer operon [1, 84], a defense only available in prokaryotes that have acquired the mer locus through horizontal gene transfer.
Releasing protein-bound iron
Mercury’s high affinity for thiol ligands presents a challenge to enzymes that use iron-sulfur clusters [50, 85]. E. coli’s labile iron pool is approximately 100 µM [50] and exposure to HgCl2 at a 3.5-fold molar excess over cellular thiols can release as much as 80 % of the labile iron pool to the free state. Our EPR data for free iron in the unexposed control culture (Fig. 3) were within range of other published data from E. coli grown in other minimal media [22, 33] and were very reproducible.
Exposure to organic oxidizing compounds only resulted in marginal increases in free iron, indicating that cellular pathways that provide protection and repair from ROS are relatively effective at combating exposure to these chemicals. Merthiolate exposure resulted in a small but not significant increase in free iron. PMA’s effectiveness in blocking thiols was reflected in still greater release of free iron; however, inorganic Hg(II) was most effective in releasing Fe. It is important to note that 16 µM inorganic HgCl2 resulted in a greater increase in free iron, relative to 40 µM PMA, even though Table 1 shows that more mercury was present in the cell in the PMA exposure. This difference shows that the degree of damage induced is not the same for both inorganic and organic mercury, i.e. they are qualitatively different in their in vivo biochemical effects. Similar ligand preferences of mercury and ferrous iron likely increase mercury’s effectiveness in disrupting labile iron centers. Inorganic mercury was only observed in peptides with multiple proximal cysteines (Table S7), a motif common among Fe-S cluster proteins [86]. In addition to direct damage caused by mercury binding, disruption of iron homeostasis can lead to oxidative stress, resulting in further damage to proteins and DNA by ROS [83].
Disrupting electrolyte balance
Mercury also disrupted the cellular electrolyte balance likely via the KefC antiporter, which is inactive when bound to reduced glutathione. Glutathione-S-conjugates induce a conformational change in KefC that activates efflux of K+ [57]. Bis-coordination of GSH by Hg will deplete the free GSH pool and our results are the first to suggest that mercurials can activate KefC. The resulting rapid efflux of K+ ions and concomitant import of H+ ions by KefC is believed to protect the cell [58] by decreasing the cellular pH, so as to protonate potential binding sites for electrophiles. The increase in Na+ content may result from activation of sodium uptake by NhaA (a Na+/H+ antiporter) under low intracellular pH conditions leading to restoration of the electrolyte balance [59–61].
Effects of mercury compounds on specific proteins
Given our observation of Hg-provoked intracellular Fe(III) release, we examined closely the effects of mercurial exposure on the subset of Fe-binding proteins observed in our larger proteomics study. Proteomics analysis detected 20 Fe-binding proteins modified by mercury (Table S7) and in many cases the mercury-binding site was a cysteine residue that coordinates an Fe-S cluster and would not be easily repaired without the proteins of the mer operon [84, 87]. The mercury vulnerable iron-binding proteins we observed play critical roles ranging from Fe-S cluster assembly (IscA and IscU), to redox defense (Fdx, MsrB, SodB, YggX) and amino acid biosynthesis (GltB and GltD).
Mercurials also formed adducts with proteins that bind other transition metals, primarily zinc (Table S8), which like mercury is a group 12, soft, divalent metal. However, since Zn is spectroscopically silent and Zn fluorescent probes do not penetrate bacterial cells, we could not assess how much Zn was freed from its natural protein sites by exposure to mercurials. Notably, ribosomal protein L31, which is implicated in Zn storage [88], was observed with mercurial adducts.
Conclusions
Inorganic mercury in short term assays is more effective at blocking total and protein thiols than PMA, and both compounds are more effective in such blockage than merthiolate, likely due to its inefficient uptake. All three mercurials disturb the electrolyte balance, but none of them provokes bulk loss of the alkaline earth metal, Mg, nor of 6 essential transition metals. When Hg(II) exceeds the available cellular thiol pool, non-thiol cellular targets such as nitrogen ligands in nucleotide bases and protein histidine and amine groups form complexes with it, but this is not seen with PMA. Characteristically cysteine-rich Fe-binding and Zn-binding proteins formed mass spectrometry stable adducts with organic and inorganic mercurials. Although each mercurial caused similar thiol blockage in 30 min exposures, inorganic Hg(II) was strikingly more effective in freeing protein-bound iron and bound most stably to peptides capable of chelating it with two or more cysteines. Thus, there are both quantitative and qualitative differences in the biochemical effects of these mercurials in living cells. Our novel findings on cellular Hg(II) biochemistry emphasize that the inorganic forms Hg(II) and Hg(0), from which it is derived, can damage cells as effectively as the organomercurials, which are commonly referred to by non-specialists as the most toxic forms of Hg. These are the first in vivo observations in a well defined model organism of the biochemical differences of inorganic and organic mercury exposure that contribute to their distinct toxicological profiles in bacteria and likely also higher organisms. It is also the first to assess the use of mass spectrometry proteomics for qualitative measurement of stable protein-mercurial adducts.
References
Barkay T, Miller SM, Summers AO (2003) FEMS Microbiol Rev 27:355–384
Mason RP, Fitzgerald WF, Morel FMM (1994) Geochim Cosmochim Acta 58:3191–3198
Norn S, Permin H, Kruse E, Kruse PR (2008) Dan Medicin Arbog 36:21–40
Crinnion WJ (2000) Altern Med Rev 5:209–223
Richardson GM, Wilson R, Allard D, Purtill C, Douma S, Graviere J (2011) Sci Total Environ 409:4257–4268
Malm O (1998) Environ Res 77:73–78
Bakir F, Damluji SF, Amin-Zaki L, Murtadha M, Khalidi A, Al-Rawi NY, Tikriti S, Dahahir HI, Clarkson TW, Smith JC, Doherty RA (1973) Science 181:230–241
Yorifuji T, Tsuda T, Takao S, Harada M (2008) Epidemiology 19:3–9
Davidson PW, Myers GJ, Weiss B (2004) Pediatrics 113:1023–1029
Clarkson TW, Magos L (2006) Crit Rev Toxicol 36:609–662
Cheesman BV, Arnold AP, Rabenstein DL (1988) J Am Chem Soc 110:6359–6364
Oram PD, Fang X, Fernando Q, Letkeman P, Letkeman D (1996) Chem Res Toxicol 9:709–712
Valko M, Morris H, Cronin MT (2005) Curr Med Chem 12:1161–1208
Schafer FQ, Buettner GR (2001) Free Radic Biol Med 30:1191–1212
Miseta A, Csutora P (2000) Mol Biol Evol 17:1232–1239
Carvalho CM, Chew EH, Hashemy SI, Lu J, Holmgren A (2008) J Biol Chem 283:11913–11923
O’Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J (1993) J Biol Chem 268:9063–9070
Imesch E, Moosmayer M, Anner BM (1992) Am J Physiol 262:F837–F842
Soskine M, Steiner-Mordoch S, Schuldiner S (2002) Proc Natl Acad Sci USA 99:12043–12048
Khan MA, Wang F (2009) Environ Toxicol Chem 28:1567–1577
Gladyshev VN, Kryukov GV (2001) BioFactors 14:87–92
Finney LA, O’Halloran TV (2003) Science 300:931–936
Helbig K, Bleuel C, Krauss GJ, Nies DH (2008) J Bacteriol 190:5431–5438
Ercal N, Gurer-Orhan H, Aykin-Burns N (2001) Curr Top Med Chem 1:529–539
Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM (2008) J Biol Inorg Chem 13:1205–1218
Waldron KJ, Rutherford JC, Ford D, Robinson NJ (2009) Nature 460:823–830
Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL II, Jenney FE Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MW (2010) Nature 466:779–782
Polacco BJ, Purvine SO, Zink EM, Lavoie SP, Lipton MS, Summers AO, Miller SM (2011) Mol Cell Proteomics 10(M110):004853
Neidhardt FC, Bloch PL, Smith DF (1974) J Bacteriol 119:736–747
Bradford MM (1976) Anal Biochem 72:248–254
Cayley S, Record MT Jr (2003) Biochemistry 42:12596–12609
Ellman GL (1959) Arch Biochem Biophys 82:70–77
Woodmansee AN, Imlay JA (2002) Methods Enzymol 349:3–9
Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O’Donovan C (2015) Nucleic Acids Res 43:D1057–D1063
Kim S, Gupta N, Pevzner PA (2008) J Proteome Res 7:3354–3363
Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Anal Chem 74:5383–5392
Scott RA (2000) Physical methods in bioinorganic chemistry—spectroscopy and magnetism. University Science Books, Sausalito, pp 465–504
George GN, Garrett RM, Prince RC, Rajagopalan KV (1996) J Am Chem Soc 118:8588–8592
Ankudinov AL, Bouldin CE, Rehr JJ, Sims J, Hung H (2002) Phys Rev B 65
Mustre de Leon J, Rehr JJ, Zabinsky SI, Albers RC (1991) Phys Rev B Condens Matter 44:4146–4156
Cosper NJ, Stalhandske CM, Saari RE, Hausinger RP, Scott RA (1999) J Biol Inorg Chem 4:122–129
Tyagarajan K, Pretzer E, Wiktorowicz JE (2003) Electrophoresis 24:2348–2358
Fruchter RG, Crestfield AM (1967) J Biol Chem 242:5807–5812
Boja ES, Fales HM (2001) Anal Chem 73:3576–3582
Cotner RC, Clagett CO (1973) Anal Biochem 54:170–177
Basinger MA, Casas J, Jones MM, Weaver AD, Weinstein NH (1981) J Inorg Nucl Chem 43:1419–1425
Khokhlova A, Chernikova G, Shishin L (1982). Inst obs neorg khimii im ns kurnakova leninski prospekt 31, 71 Moscow, Russia, pp 2976–2978
Powell KJ, Brown PL, Byrne RH, Gajda T, Hefter G, Sjoberg S, Wanner H (2005) IUPAC. Pure Appl Chem 77:739–800
Johnson DC, Dean DR, Smith AD, Johnson MK (2005) Annu Rev Biochem 74:247–281
Keyer K, Imlay JA (1997) J Biol Chem 272:27652–27659
Lafrance-Vanasse J, Lefebvre M, Di Lello P, Sygusch J, Omichinski JG (2009) J Biol Chem 284:938–944
Parks JM, Guo H, Momany C, Liang L, Miller SM, Summers AO, Smith JC (2009) J Am Chem Soc 131:13278–13285
Xu FF, Imlay JA (2012) Appl Environ Microbiol 78:3614–3621
Stricks W, Kolthoff IM (1953) J Am Chem Soc 75:5673–5681
Güzeloğlu Ş, Yalçın G, Pekin M (1998) J Organomet Chem 568:143–147
McClintock CS, Parks JM, Bern M, Ghattyvenkatakrishna PK, Hettich RL (2013) J Proteome Res 12:3307–3316
Roosild TP, Castronovo S, Healy J, Miller S, Pliotas C, Rasmussen T, Bartlett W, Conway SJ, Booth IR (2010) Proc Natl Acad Sci USA 107:19784–19789
Ferguson GP (1999) Trends Microbiol 7:242–247
Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H (2005) Nature 435:1197–1202
Padan E (2011) Compr Physiol 1:1711–1719
Taglicht D, Padan E, Schuldiner S (1991) J Biol Chem 266:11289–11294
Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D (2005) Arch Microbiol 183:9–18
Zheng M, Doan B, Schneider TD, Storz G (1999) J Bacteriol 181:4639–4643
Nies DH (2003) FEMS Microbiol Rev 27:313–339
Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, Tanaka Y, Kondo Y, Sawa R, Fujimoto T, Machinami T, Ono A (2006) J Am Chem Soc 128:2172–2173
Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kojima C, Ono A (2007) J Am Chem Soc 129:244–245
Brooks P, Davidson N (1960) J Am Chem Soc 82:2118–2123
Bligh EG, Dyer WJ (1959) Can J Biochem Physiol 37:911–917
Summers AO, Wireman J, Vimy MJ, Lorscheider FL, Marshall B, Levy SB, Bennett S, Billard L (1993) Antimicrob Agents Chemother 37:825–834
Rietschel RL, Wilson LA (1982) Arch Dermatol 118:147–149
Tosti A, Tosti G (1988) Contact Dermatitis 18:268–273
Freed LF (1948) S Afr Med J 22:223–229
Weed LE, Ecker EE (1931) J Infect Dis 49:440–449
Ball LK, Ball R, Pratt RD (2001) Pediatrics 107:1147–1154
WHO (2002) Wkly Epidemiol Rec 77:305–316
Gutknecht J (1981) J Membr Biol 61:61–66
Barkay T, Gillman M, Turner RR (1997) Appl Environ Microbiol 63:4267–4271
Owens RA, Hartman PE (1986) J Bacteriol 168:109–114
Eser M, Masip L, Kadokura H, Georgiou G, Beckwith J (2009) Proc Natl Acad Sci USA 106:1572–1577
Ndu U, Mason RP, Zhang H, Lin S, Visscher PT (2012) Appl Environ Microbiol 78:7276–7282
Mah V, Jalilehvand F (2008) J Biol Inorg Chem 13:541–553
Ravichandran M (2004) Chemosphere 55:319–331
Imlay JA (2013) Nat Rev Microbiol 11:443–454
Ledwidge R, Patel B, Dong A, Fiedler D, Falkowski M, Zelikova J, Summers AO, Pai EF, Miller SM (2005) Biochemistry 44:11402–11416
Jung YS, Yu L, Golbeck JH (1995) Photosynth Res 46:249–255
Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F (2013) Biochim Biophys Acta 1827:455–469
Hong B, Nauss R, Harwood IM, Miller SM (2010) Biochemistry 49:8187–8196
Gabriel SE, Helmann JD (2009) J Bacteriol 191:6116–6122
Acknowledgments
We thank Mary Lipton, Erika Zink, and Samuel Purvine (all of the DOE Pacific Northwest National Laboratory) for chemical and biophysical acquisition and SEQUEST analysis of the proteomic data, Tejas Chaudhari and Sagar Tarkhadkar (Department of Computer Sciences, Univ. of Georgia) for assistance with database development and management, and Graham George (University of Saskatchewan and the Canadian Light Source) for mercuric bromide EXAFS data collection. This work was supported by DOE awards ER64408 and ER65286 to AOS and ER64409 and ER65195 to SMM and NIH award GM62524 to MKJ.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
LaVoie, S.P., Mapolelo, D.T., Cowart, D.M. et al. Organic and inorganic mercurials have distinct effects on cellular thiols, metal homeostasis, and Fe-binding proteins in Escherichia coli . J Biol Inorg Chem 20, 1239–1251 (2015). https://doi.org/10.1007/s00775-015-1303-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-015-1303-1