Abstract
The purpose of this study was to identify relationships between single nucleotide polymorphisms (SNPs) in the genes of the Wnt pathway and bone mineral density (BMD) of postmenopausal women. We chose this pathway due to its importance in bone metabolism that was underlined in several studies. DNA samples of 932 Hungarian postmenopausal women were studied. First, their BMD values at different sites (spine, total hip) were measured, using a Lunar Prodigy DXA scanner. Thereafter, T-score values and the patients’ body mass indices (BMIs) were calculated, while information about the fracture history of the sample population was also collected. We genotyped nine SNPs of the following three genes: LRP5, GPR177, and SP7, using a Sequenom MassARRAY Analyzer 4 instrument. The genomic DNA samples used for genotyping were extracted from the buccal mucosa of the subjects. Statistical analyses were carried out using the SPSS 21 and R package. The results of this analysis showed a significant association between SNP rs4988300 of the LRP5 gene and total hip BMD values. We could not reveal any associations between the markers of GPR177, SP7, and bone phenotypes. We found no effect of these genotypes on fracture risk. We could demonstrate a significant gene–gene interaction between two SNPs of LRP5 (rs4988300 and rs634008, p = 0.009) which was lost after Bonferroni correction. We could firmly demonstrate a significant association between rs4988300 of the LRP5 gene and bone density of the hip on the largest homogeneous postmenopausal study group analyzed to date. Our finding corroborates the relationship between LRP5 genotype and bone phenotype in postmenopausal women, however, the complete mechanism of this relationship requires further investigations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoporosis is a complex disease with a strong genetic background [1]. The genetic effects are mediated through a wide variety of genes [2]. Genetic effects were observed in all phases of bone metabolism, in bone formation as well as in bone resorption. The receptor activator of nuclear factor kappa-B (RANK)/receptor activator of the nuclear factor kappa-B ligand (RANKL) pathway is a crucial element in bone resorption [3]. Genome-wide association studies (GWAS) studies showed that single nucleotide polymorphisms (SNPs) in the genes of the RANK/RANKL pathway were associated with bone mass and fracture risks [4, 5].
The Wnt pathway plays an important role in bone metabolism, especially bone formation, and its alterations are associated with osteoporosis [6–11]. Members of this pathway are in close relationship with bone development and bone density. The most investigated factor of the Wnt pathway in osteoblast differentiation is the LRP5/6 [12]. The gain of function mutations of LRP5/6 result in high bone mass or osteopetrosis, and the loss of function mutations lead to osteoporosis-pseudoglioma syndrome [13–15]. So far, all GWAS studies concentrating on LRP5/6 gene have found a relationship between its genotypes and osteoporosis [16, 17].
GPR177 is a 7-transmembrane protein with an effect on the secretion of Wnt proteins; it facilitates the excretion of Wnts from the Golgi to the extracellular space [18]. Loss of GPR177 leads to disturbed axial differentiation in mice. Therefore, GPR177-null mice show severe impairment in skeletal development, which suggests that GPR177 plays an important role in the Wnt pathway [19]. Furthermore, a recent GWAS study showed that GPR177 affects bone mass in humans [20].
SP7 (also known as osterix) is a zinc-finger containing a transcription factor of bone formation and osteoblast differentiation. The SP7 and Hypoxia Inducible Factor-1alpha (HIF1A) have been demonstrated to synergistically inhibit the Wnt pathway [21]. TheSP7 is important in shifting mesodermal cells away from the cartilage cell line towards osteoblast lineage [22]. The SP7 gene expression in circulating mesenchymal stem cells was decreased in osteoporotic patients compared to healthy individuals [23]. In the Sister Study, the SNPs of the SP7 gene appeared to be associated with bone mineral density (BMD) in African–American women [16].
Based on these previous studies, it seemed likely that the polymorphisms of these genes in the WNT pathway might have an effect on bone metabolism. Therefore, we tested how the functioning SNPs of LRP5, GPR177, and SP7 genes affected BMD and fracture risk in Caucasian postmenopausal women.
Materials and methods
Patient and phenotypic information
The population in our study consisted of 932 postmenopausal, non-related women referred to Hungarian osteoporosis centers. Patients were considered postmenopausal if they were older than 40 and there was no menstruation period within 1 year prior to our study. Secondary osteoporosis was excluded. We collected data on patients’ age, height, and body weight to calculate BMI. Patient data can be seen in Table 1. After measuring femoral and spinal BMD using a Lunar Prodigy DXA scanner (GE Medical Systems, Madison, Wisconsin, USA), we calculated femoral and spinal T-scores based on the Hungarian reference range. We sorted subjects into different diagnostic subgroups (osteoporosis, osteopenia, and normal bone status) based on the WHO criteria. Patients with a T-score less than −2.5 at any site were considered osteoporotic, patients with a T-score value between −2.5 and −1.0 at any site are osteopeniac, and subjects with T-score values higher than −1.0 have normal bone status. The fracture history of a subpopulation of 385 patients, obtained from their medical history, was also utilized. Non-vertebral osteoporotic fracture was defined as a low-trauma fracture after the age of 40 years excluding the fractures of the face, skull, fingers, toes, and spine. Vertebral compression fractures were not investigated in this study. Written informed consent was obtained from all participants. The study was approved by the Science and Research Ethics Committee of the Medical Science Council, Hungary.
SNP selection
We used publicly available online databases (http://genome.ucsc.edu/, http://www.genome.gov/gwastudies/, and http://www.ncbi.nlm.nih.gov/omim) to select SNPs of the following genes: LRP5, GPR177, SP7.
Nine SNPs were chosen based on their function or previous appearance in GWAS studies. On the basis of the first criteria, missense variations were chosen which cause a change in the amino acid sequence of the encoded protein. Regarding the second criteria, we chose SNPs that have shown a p value less than or equal to 5 × 10−8.
Genotyping
We collected genomic DNA from the patients’ buccal mucosa, brushing off the superficial layer of cells lining the oral cavity. DNA was extracted using a High Pure PCR Template Purification kit (Roche Diagnostics, GmbH, Mannheim, Germany), and genotyping was performed on a Sequenom MassARRAY Analyzer 4 (Sequenom, San Diego, CA, USA).
Statistical analysis
Genotyping data and fracture risk
To determine the relationship between genotypes and fracture risk, we created contingency tables containing the genotypes and fracture history. To test the statistical significance of the results, we applied Pearson’s chi-squared test.
Genotyping data and bone parameters
The subjects were assigned to different groups based on the genotyping results, meaning that three groups (homozygote recessive, homozygote dominant, and heterozygote) were created for each SNP. The genotype groups were tested for normality using Shapiro–Wilk’s test and homogeneity of variances by Levene’s test. We utilized analysis of covariance (ANCOVA) to test the different genotypes against each other, and the Bonferroni method was used to adjust for multiple testing. We adjusted BMD values for age and BMI. All tests were performed using SPSS 21 (SPSS Inc., Chicago, IL, USA). Linkage disequilibrium plots based on our own data were created using HaploView 4.0 (Broad Institute, Cambridge, MA, USA). Interactions of different genes on phenotype were calculated with SNPassoc [24], haplotype analyses were carried out using haplo.stats. Both of these are R packages (R Foundation for Statistical Computing, Vienna, Austria). We chose an alpha value of 0.05.
Results
Basic statistics of subject group and pre-analysis
Basic statistical characteristics of the sample group are detailed in Table 1. Forty-three samples of our initial sample size of 932 did not contain enough DNA for genotyping, and they were excluded from further analysis. Published results here (Table 2) are based on the remaining 889 women. Table 2 contains the bone parameters of our subjects in the different study groups.
Genotyping results
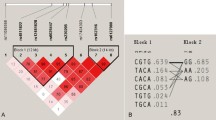
General data about genotyping are presented in Table 3. All of the genotyped SNPs followed the Hardy–Weinberg equilibrium. Call rate was at least 98.54 % for all SNPs. One SNP turned out to be monomorphic in our population (SP7 rs191240606), even though it was polymorphic in all populations of the 1000 genomes project. The SNP is not present in the HapMap database. Genotyped SNPs in GPR177 formed two haplotype blocks, while in LRP5 we could create one haplotype block (Fig. 1).
Influence of LRP5 SNPs on bone mass and fracture risk
We found that rs4988300 and rs634008 had an association with total hip BMD. The association between rs4988300 and total hip BMD remained significant after Bonferroni correction (p = 0.004). Heterozygotes of this SNP have a significantly higher total hip BMD than homozygotes. Our data showed an association between rs599083, rs634008, and total hip T-score, respectively (p 1 = 0.040, p 2 = 0.034), but the significance disappeared after Bonferroni correction. We found no correlation between spine BMD and the tested genotypes. Furthermore, we observed no interaction between the tested genotypes and the incidence of osteoporotic fractures at any sites.
Haplotype analysis and gene–gene interactions
A gene–gene interaction plot was drawn using SNPassoc (Fig. 2). The gene–gene interaction plot suggested an interaction between two SNPs of LRP5 (rs4988300 and rs634008, p = 0.009), which was lost after Bonferroni correction. No other interactions were found between the studied SNPs. Using the haplo.score function of haplo.stats, we found no significant change in total hip T-score or total hip BMD. Additional information about the haplotype analysis can be found in Table 4.
Influence of GPR177and SP7 SNPs on bone mass and fracture risk
We were unable to find any associations between SNPs of these two genes and BMD of our subjects. We observed no association between the tested genotypes and the incidence of osteoporotic fractures at any site. (No data shown).
A table with the adjusted BMD and T-score values for all genotypes of the selected SNPs is provided as supplementary material.
Discussion
In this study, we investigated the relationship between the candidate genetic variations of the Wnt pathway and bone density/fracture rate. We could firmly demonstrate a significant association between rs4988300 of the LRP5 gene and bone density of the hip on the largest homogeneous postmenopausal study group analyzed to date.
LRP5 is a co-receptor of the Wnt pathway; several loci of this gene were described as potential factors in osteoporosis by GWAS studies [25]. Loss of function mutations in this gene lead to osteoporosis-pseudoglioma syndrome [26, 27]. Gain of function mutations result in high bone density [28]. A strong effect of LRP5 gene SNPs on BMD and fracture risk has been established [13–15, 29, 30]. A report by Xiong et al. [31] concluded that four genes (LRP5, DBP, CYP17, and RANK) showed highly suggestive associations with spine BMD. They found that rs4988300 showed an association with spine osteoporosis (OP) and rs634008 was associated with spine hip and ultradistal radius OP. However, these findings lost significance after correction for the number of haplotypes in the haplotype-based association test. Rs4988300 showed a statistically significant association with total hip BMD in our samples even after Bonferroni correction. This finding is in accordance with the previous results of Kiel et al. [32]. Our study cohort was stratified for gender, this way we could eliminate the difficulties caused by the different pathogenesis in men and women. Our study group was bigger for osteoporotic women than that of Kiel et al. They studied 646 postmenopausal women; we had genotyping information on 889 subjects. We also had a subgroup of women with detailed fracture history. In the study of Kiel et al., genotypes of rs4988300 were associated with femur shaft section modulus in males. Based on this data, we tested if the genotypes influenced fracture risk. We found no association between these traits; however, there are other factors besides BMD that influence fracture risk. These include cortical thickness of bone and bone microarchitecture [33]. Differences in these bone parameters might explain why the genotypes did not associate with fracture history.
We could not find evidence for the role of the other two factors of the Wnt pathway, SP7 and GPR177. This does not rule out the association with BMD; however, the effect seems to be too small to be identifiable in our study population of osteoporotic women. GPR177 is a glycoprotein, which mediates secretion signals for the Wnt pathway [34]. In rodents GPR177 is expressed even in adulthood, its presence is essential for healthy organogenesis [35]. It is also essential for adequate antero-posterior development. Fu et al. [36] hypothesized a reciprocal interaction between GPR177 and Wnt to regulate the levels of both factors. GPR177 level elevates as it is a target of Wnt, as a result GPR177 increases Wnt signaling by promoting Wnt excretion from the Golgi. Kumar et al. [6] found no association of BMD with the SNPs (rs983034, rs3748705, rs2566755, and rs2820475) of GPR177 gene on groups of postmenopausal and young women. For the same SNPs (rs983034 and rs2566755), we could not find an association with BMD in our study population either. In contrast, Roshandel et al. [37] showed a significant association between spinal BMD of men and GPR177 gene SNP rs1430742 in 2,359 participants of the European Male Ageing Study (EMAS).We also included this SNP in our study; however, we found no statistically significant association between this trait and BMD in our study group. The reason for this difference might be explained by the different genders included in the two studies. Rs983034 and rs3748704 code a missense variation; however, rs983034 did not cause a detectable change of bone status in our study group. Even though previous GWAS studies (studying rs2566755 and rs1430742) could demonstrate a slight effect on BMD, our results could not corroborate this effect.
SP7 has a known effect on bone development; it is involved in embryogenesis, and has a role in bone reparation too [38]. SP7 has a decreased expression in the circulating mesenchymal stem cells of osteoporotic patients [23]. Another study showed no association between SP7 genotypes and BMD [39]. In our study, we could not find evidence for any relationship between rs2016266 and BMD or fracture history. Rs191240606 turned out to be monomorphic despite the fact that this SNP is not described as monomorphic in any population of the 1000 genomes project (http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/). There is no data collected from Eastern European populations in the 1000 genomes project, thus, a putative difference between the Hungarian population and CEU (US residents of northern and western European ancestry) population might explain this finding.
As with every study, ours has its strengths and limitations. The advantages are that we had a relatively large, genetically homogenous study group. The main disadvantages include a low number of cases in the fracture history subgroup, a low number of SNPs per gene, a small cohort size for haplotype analyses, and we did not examine the biological background associated with the genotypes. Despite these pitfalls, our study further strengthened the close relationship of LRP5 genotypes and bone density as well as provided more insights into the cause of the so far often contradictory results by clarifying them in a rather confounding factors-free study environment.
References
Zaidi M (2007) Skeletal remodeling in health and disease. Nat Med 13:791–801
Hsu YH, Kiel DP (2012) Clinical review: genome-wide association studies of skeletal phenotypes: what we have learned and where we are headed. J Clin Endocrinol Metab 97:E1958–E1977
Paternoster L, Lorentzon M, Vandenput L et al (2010) Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genet 6:e1001217
Takacs I, Lazary A, Kosa JP et al (2010) Allelic variations of RANKL/OPG signaling system are related to bone mineral density and in vivo gene expression. Eur J Endocrinol 162:423–431
Lazary A, Kosa JP, Tobias B et al (2008) Single nucleotide polymorphisms in new candidate genes are associated with bone mineral density and fracture risk. Eur J Endocrinol 159:187–196
Kumar J, Swanberg M, McGuigan F, Callreus M, Gerdhem P, Akesson K (2011) LRP4 association to bone properties and fracture and interaction with genes in the Wnt- and BMP signaling pathways. Bone 49:343–348
Li X, Grisanti M, Fan W et al (2011) Dickkopf-1 regulates bone formation in young growing rodents and upon traumatic injury. J Bone Miner Res 26:2610–2621
Wend P, Wend K, Krum SA, Miranda-Carboni GA (2012) The role of WNT10B in physiology and disease. Acta Physiol (Oxf) 204:34–51
Baron R, Kneissel M (2013) WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 19:179–192
Maeda K, Takahashi N, Kobayashi Y (2013) Roles of Wnt signals in bone resorption during physiological and pathological states. J Mol Med (Berl) 91:15–23
Regard JB, Zhong Z, Williams BO, Yang Y (2012) Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb Perspect Biol 4:a007997
Day TF, Guo X, Garrett-Beal L, Yang Y (2005) Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell 8:739–750
Gong Y, Slee RB, Fukai N et al (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107:513–523
He X, Semenov M, Tamai K, Zeng X (2004) LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development 131:1663–1677
Yadav VK, Ryu JH, Suda N et al (2008) Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 135:825–837
Ichikawa S, Koller DL, Padgett LR, Lai D, Hui SL, Peacock M, Foroud T, Econs MJ (2010) Replication of previous genome-wide association studies of bone mineral density in premenopausal American women. J Bone Miner Res 25:1821–1829
Estrada K, Styrkarsdottir U, Evangelou E et al (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44:491–501
Bartscherer K, Pelte N, Ingelfinger D, Boutros M (2006) Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125:523–533
Maruyama T, Jiang M, Hsu W (2013) Gpr177, a novel locus for bone mineral density and osteoporosis, regulates osteogenesis and chondrogenesis in skeletal development. J Bone Miner Res 28:1150–1159
Rivadeneira F, Styrkársdottir U, Estrada K et al (2009) Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet 41:1199–1206
Chen D, Li Y, Zhou Z, Xing Y, Zhong Y, Zou X, Tian W, Zhang C (2012) Synergistic inhibition of Wnt pathway by HIF-1alpha and osteoblast-specific transcription factor osterix (Osx) in osteoblasts. PLoS ONE 7:e52948
Kobayashi T, Kronenberg H (2005) Minireview: transcriptional regulation in development of bone. Endocrinology 146:1012–1017
Dalle Carbonare L, Valenti MT, Zanatta M, Donatelli L, Lo Cascio V (2009) Circulating mesenchymal stem cells with abnormal osteogenic differentiation in patients with osteoporosis. Arthritis Rheum 60:3356–3365
Gonzalez JR, Armengol L, Sole X, Guino E, Mercader JM, Estivill X, Moreno V (2007) SNPassoc: an R package to perform whole genome association studies. Bioinformatics 23:644–645
Riancho JA, Olmos JM, Pineda B et al (2011) Wnt receptors, bone mass, and fractures: gene-wide association analysis of LRP5 and LRP6 polymorphisms with replication. Eur J Endocrinol 164:123–131
Tuysuz B, Bursali A, Alp Z, Suyugul N, Laine CM, Makitie O (2012) Osteoporosis-pseudoglioma syndrome: three novel mutations in the LRP5 gene and response to bisphosphonate treatment. Horm Res Paediatr 77:115–120
Laine CM, Chung BD, Susic M et al (2011) Novel mutations affecting LRP5 splicing in patients with osteoporosis-pseudoglioma syndrome (OPPG). Eur J Hum Genet 19:875–881
Boyden LM, Mao J, Belsky J et al (2002) High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 346:1513–1521
Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann C (2005) Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell 8:727–738
Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F (2005) Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 132:49–60
Xiong DH, Shen H, Zhao LJ et al (2006) Robust and comprehensive analysis of 20 osteoporosis candidate genes by very high-density single-nucleotide polymorphism screen among 405 white nuclear families identified significant association and gene-gene interaction. J Bone Miner Res 21:1678–1695
Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D (2007) Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet 8(Suppl 1):S14
Cefalu CA (2004) Is bone mineral density predictive of fracture risk reduction? Curr Med Res Opin 20:341–349
Yu HM, Jin Y, Fu J, Hsu W (2010) Expression of Gpr177, a Wnt trafficking regulator, in mouse embryogenesis. Dev Dyn 239:2102–2109
Jin J, Morse M, Frey C, Petko J, Levenson R (2010) Expression of GPR177 (Wntless/Evi/Sprinter), a highly conserved Wnt-transport protein, in rat tissues, zebrafish embryos, and cultured human cells. Dev Dyn 239:2426–2434
Fu J, Jiang M, Mirando AJ, Yu HM, Hsu W (2009) Reciprocal regulation of Wnt and Gpr177/mouse Wntless is required for embryonic axis formation. Proc Natl Acad Sci USA 106:18598–18603
Roshandel D, Thomson W, Pye SR et al (2011) Polymorphisms in genes involved in the NF-kappaB signalling pathway are associated with bone mineral density, geometry and turnover in men. PLoS One 6:e28031
Chauveau C, Broux O, Delecourt C, Hardouin P, Jeanfils J, Devedjian JC (2008) Gene expression in normotopic and heterotopic human bone: increased level of SP7 mRNA in pathological tissue. Mol Cell Biochem 318:81–87
Mendez JP, Rojano-Mejia D, Coral-Vazquez RM et al (2013) Impact of genetic variants of IL-6, IL6R, LRP5, ESR1 and SP7 genes on bone mineral density in postmenopausal Mexican-Mestizo women with obesity. Gene 528:216–220
Conflict of interest
All authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Horváth, P., Balla, B., Kósa, J.P. et al. Strong effect of SNP rs4988300 of the LRP5 gene on bone phenotype of Caucasian postmenopausal women. J Bone Miner Metab 34, 79–85 (2016). https://doi.org/10.1007/s00774-014-0645-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00774-014-0645-z