Abstract
The free d-amino acid, d-aspartate, is abundant in the embryonic brain but significantly decreases after birth. Besides its intracellular occurrence, d-aspartate is also present at extracellular level and acts as an endogenous agonist for NMDA and mGlu5 receptors. These findings suggest that d-aspartate is a candidate signaling molecule involved in neural development, influencing brain morphology and behaviors at adulthood. To address this issue, we generated a knockin mouse model in which the enzyme regulating d-aspartate catabolism, d-aspartate oxidase (DDO), is expressed starting from the zygotic stage, to enable the removal of d-aspartate in prenatal and postnatal life. In line with our strategy, we found a severe depletion of cerebral d-aspartate levels (up to 95%), since the early stages of mouse prenatal life. Despite the loss of d-aspartate content, Ddo knockin mice are viable, fertile, and show normal gross brain morphology at adulthood. Interestingly, early d-aspartate depletion is associated with a selective increase in the number of parvalbumin-positive interneurons in the prefrontal cortex and also with improved memory performance in Ddo knockin mice. In conclusion, the present data indicate for the first time a biological significance of precocious d-aspartate in regulating mouse brain formation and function at adulthood.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
d-Aspartate (d-Asp) and d-serine (d-Ser) are the only free d-amino acids present in substantial amounts in the mammalian brain. Over the past decades, several studies in rodents and humans revealed that d-Asp has a transient emergence in the brain during embryonic life and decreases significantly after birth (Dunlop et al. 1986; Hashimoto et al. 1993, 1995; Neidle and Dunlop 1990; Sakai et al. 1998; Wolosker et al. 2000; Punzo et al. 2016). In particular, high-performance liquid chromatography (HPLC) analysis revealed that d-Asp content is exceptionally high in the human prefrontal cortex (PFC) during early gestational stages, when it is even more abundant than the corresponding l-enantiomer (Hashimoto et al. 1993). Interestingly, the intracellular distribution of d-Asp is regulated developmentally and follows the functional activity of neuroblasts. Indeed, in the rat forebrain, d-Asp is localized in the cytoplasm of migrating neuroblasts at embryonic day 12 (E12); after the migration is complete and a distinct layer is established, d-Asp is redistributed to axonal processes (Sakai et al. 1998). Although the transient presence of d-Asp in the brain suggests the existence of a biosynthetic pathway for its production, a selective d-Asp-synthesizing enzyme has not yet been identified in mammals (Horio et al. 2013; Ito et al. 2016; Tanaka-Hayashi et al. 2014). On the other hand, it is well established that the peroxisomal enzyme, d-aspartate oxidase (DDO, or DASPO EC 1.4.3.1), is responsible for d-Asp catabolism (D'Aniello et al. 1993; Katane and Homma 2010; Van Veldhoven et al. 1991; Molla et al. 2020) and for the drastic postnatal depletion of cerebral d-Asp content (Punzo et al. 2016; Van Veldhoven et al. 1991).
Pharmacological studies have shown that d-Asp binds to (Fagg and Matus 1984; Olverman et al. 1988) and activates (Errico et al. 2008a, b, 2011a, b) the NMDA subclass of glutamate receptors (NMDAR), due to its affinity for the glutamate site of GluN2A-D subunits (Errico et al. 2011a). Further evidence has also suggested that d-Asp acts as an endogenous agonist of the metabotropic glutamate receptor, mGluR5 (Molinaro et al. 2010). Moreover, d-Asp is able to trigger the release of glutamate in specific brain regions (Cristino et al. 2015; Sacchi et al. 2017) through the presynaptic activation of NMDA, AMPA, and mGlu5 receptors (Sacchi et al. 2017). The abovementioned pharmacological findings and the presence of d-Asp within extracellular space documented in the mouse brain (Punzo et al. 2016; Sacchi et al. 2017) suggest that this d-amino acid acts as an endogenous signaling molecule within glutamatergic neurotransmission system.
Neuropharmacological characterization of animal models with abnormally high cerebral levels of d-Asp, due to targeted deletion of the Ddo gene (Ddo−/−) or exogenous administration of d-Asp (Errico et al. 2006, 2008a; Kitamura et al. 2018), is in agreement with this hypothesis. Indeed, both Ddo−/−-and d-Asp-treated animals showed increased NMDAR-dependent long-term potentiation (LTP), augmented dendritic length and spine density, and improved spatial memory (Errico et al. 2008a, b, 2011a, b, 2014; Kitamura et al. 2018; Topo et al. 2010). Consistent with the preclinical observations, oral supplementation of d-Asp increased cortical trans-synaptic glutamatergic transmission and synaptic plasticity reserve also in humans, as measured by rTMS-induced LTP, intracortical facilitation, and short-interval intracortical facilitation (Nicoletti et al. 2019).
Despite these findings suggest an influence of non-physiologically high d-Asp levels on a variety of glutamatergic processes during adulthood, the biological significance of the enriched prenatal d-Asp content in the mammalian brain is so far completely unknown. To clarify this still enigmatic issue, we generated a knockin mouse model in which the expression of Ddo begins from the zygotic stage. In these mice, we evaluated the consequences of DDO-mediated embryonic d-Asp removal on a series of neurochemical, neuroanatomical, and behavioral phenotypes at adulthood.
Materials and methods
Generation of Ddo knockin mice
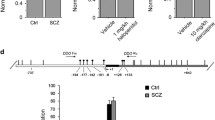
For the generation of the Ddo knockin targeting vector, a PCR-based cloning strategy was used to generate a bi-cistronic cassette, in which the IRES element simultaneously allows the expression of Ddo and LacZ; the IRES sequence was obtained from the plasmid pIRES (Clonetech) and the LacZ from the pWHERE (InvivoGen). This cassette was cloned between the attL1/attL2 sequences of the pENTR™1A vector (ThermoFisher Scientific), so that, by Invitrogen™ Gateway™ recombination cloning (ThermoFisher Scientific), it was easily moved in a destination plasmid containing the ROSA26 homology arms, the adenovirus splicing acceptor (SA), a promoter-less Neo resistance sequence flanked by FRT sites, a floxed triple polyA (pA) transcriptional stop cassette and the attR1/attR2 sequences (vector collection of the Animal Models Core Facility, AMCF, of Biogem, Ariano Irpino, Italy). The linearized targeting vector was electroporated into E14Tg4a2 embryonic stem cell line (sv129 strain) as described (Silberschmidt et al. 2011) and the G418-resistant clones were screened by long-range PCR, using the Expand™ Long Template PCR System (Sigma-Aldrich) with the primers R26sc Fw (5′-GGT CGT GTG GTT CGG TGT CTC TTT TCT GTT GG-3′) and R26sc Rev (5′-CGG CCT CGA CTC TAC GAT ACC GTC GAT CC-3′), following the manufacturer’s protocol. The correctly targeted ES cells were further analyzed by Southern blotting. In particular, the genomic DNA was digested with EcoRV and hybridized with a DIG-labeled probe (PCR DIG Probe Synthesis Kit, Sigma-Aldrich) that recognizes a portion of the 5′ homology arm, as indicated in Fig. 1. A positive ES clone was electroporated with a FLP-expressing plasmid, pCAGGS-Flpe (Gene Bridges, Heidelberg, Germany) to delete the Neo cassette by recombination between the FRT sites. One of the ES positive clones was injected into C57BL/6J blastocysts to generate chimeric mice. Chimeric founder male mice were bred with C57BL/6J females for germline transmission and the consequent generation of mice carrying the Ddo-LacZ cassette (LoxP-STOP-LoxP-Ddo-IRES-LacZ-pA; LSL-DdoLacZ) to the ROSA26 locus. R26LSL−DdoLacZ mice were genotyped by PCR using the following primers: R26 wt Fw, 5′-CTG TGG ACA GAG GAG CCA TAA C-3′; R26 wt Rev, 5′-CTG TCT CTG CCT CCA GAG TGC T-3′; LSL-DdoLacZ Rev 5′-AGT TAT CCT GAT CAC CCC TCC G-3′. The wild-type fragment (316 bp) was generated by the primers R26 wt Fw and R26 wt Rev; the targeted fragment (411 bp) was generated by the primers R26 wt Fw and LSL-DdoLacZ Rev. Amplification conditions were 95 °C for 30 s; 35 cycles: 95 °C for 30 s, 57 °C for 30 s, 72 °C for 1 min; 72 °C for 10 min. Heterozygous R26LSL−DdoLacZ mice were crossed with Hprt-Cre mice for the excision of the stop cassette. The resulting heterozygous knockin mice (R26Ddo/+) were backcrossed into the C57BL/6 J background for three generations (F3). R26Ddo/+ mice were then interbred to produce homozygous R26Ddo/Ddo animals. Knockin mice were genotyped by PCR using the following primers: R26 wt Fw, 5′-CTG TGG ACA GAG GAG CCA TAA C-3′; R26 wt Rev, 5′-CTG TCT CTG CCT CCA GAG TGC T-3′; DdoLacZ Rev 5′-TGG ACT AAA CAC CGG TGC CC-3′. The wild-type fragment (316 bp) was generated by the primers R26 wt Fw and R26 wt Rev; the targeted fragment (490 bp) was generated by the primers R26 wt Fw and DdoLacZ Rev. Amplification conditions were: 95 °C for 5 min; 35 cycles: 95 °C for 30 s, 62 °C for 30 s, 72 °C for 1 min; 72 °C for 10 min.
Generation of Ddo knockin mice. a Targeting the genomic locus of ROSA26 locus. pENTR-Ddo is the donor vector used with the Gateway™ strategy to insert the Ddo-IRES-LacZ cassette in the R26 targeting vector. The resulting construct inserts all the exogenous DNA composed of the adenovirus splicing acceptor (SA), FRT-neo-FRT, loxP-stop-loxP and Ddo-Ires-LacZ sequences in the Rosa26 genomic locus by homologous recombination. After the construct electroporation, the G418-resistant clones were screened by b PCR using the couple of primers R26scFw–R26scRev, which anneal in a region upstream the 5′ homology arm and the neo cassette, respectively. Only the correctly recombined clones amplify a ~ 3.6 Kbp long band. c The clones selected by PCR were analyzed by Southern blotting. In particular, the genomic DNA extracted from the cells was digested with EcoRV and the blotted electrophoretic run was probed with a DIG-labeled probe that recognizes a portion of the 5′ homology arm, labeling both the wild type and recombinant allele, corresponding to the ~ 11.5 and ~ 4.2 Kbp, respectively. After subsequent deletions of the neo and stop cassettes by FLP and Cre recombinases, respectively, the constitutive expression of Ddo cassette is allowed to proceed under the control of the Rosa26 promoter (R26Ddo allele). The resulting heterozygous knockin mice (R26Ddo/+) were interbred to generate homozygous mice (R26Ddo/Ddo). d The offsprings from R26Ddo/+ intercrosses were genotyped by PCR analysis of tail DNA. The bands corresponding to wild-type (316 bp) and mutant (490 bp) alleles were obtained using the couples of primers R26wtFw–R26wtRev and R26wtFw–DdoLacZRev, respectively. eR26Ddo/+ and R26Ddo/Ddo mice are undistinguishable from wild-type (R26+/+) littermates
Animals were group housed (five per cage), at a constant temperature (22 ± 1 °C) on a 12 h light/dark cycle (lights on at 7 AM) with food and water ad libitum. All the experiments in adult phase were performed on 3/4-month-old R26+/+, R26Ddo/+ and R26Ddo/Ddo male mice.
Quantitative real-time PCR
Whole brains were collected from R26+/+, R26+/Ddo, and R26Ddo/Ddo mice at different developmental stages: E16.5, E18.5, P3, P7, P14, and P90. The PFC, hippocampus, striatum, and cerebellum were dissected out. Animals were killed and the whole brain, PFC, hippocampus, striatum, and cerebellum were dissected out within 20 s on an ice-cold surface. All tissue samples were pulverized in liquid nitrogen and stored at − 80 °C for subsequent processing.
Total RNA was extracted by RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions (Querques et al. 2015). The integrity of the isolated RNAs was assessed using denaturing agarose gel electrophoresis (presence of sharp 28S, 18S, and 5S bands) and spectrophotometry (NanoDrop 2000, Thermo Scientific). Total RNA was purified to eliminate potentially contaminating genomic DNA using recombinant DNase (QIAGEN). A total of 1 μg of total RNA of each sample was reverse transcribed with Quanti Tect Reverse Transcription (QIAGEN) using oligo-dT and random primers according to the manufacturer’s instructions. qRT-PCR amplifications were performed using LightCycler 480 SYBR Green I Master (Roche Diagnostic) in a LightCycler480 Real Time thermocycler. The following protocol was used: 10 s for initial denaturation at 95 °C followed by 40 cycles consisting of 10 s at 95 °C for denaturation, 10 s at 60 °C for annealing, and 6 s for elongation at 72 °C temperature. The following primers were used for Ddo, Daao, and Sr cDNA amplification: Ddo forward, 5′-ACC ACC AGT AAT GTA GCG GC-3′ and Ddo reverse, 5′-GGT ACC GGG GTA TCT GCA C-3′; Daao forward, 5′-TTT TCT CCC GAC ACC TGG C-3 and Daao reverse, 5′-TGA ACG GGG TGA ATC GAT CT-3′; Sr forward, 5′-CCC TTG GTA GAT GCA CTG GT-3′ and Sr reverse, 5′-TCA GCA GCG TAT ACC TTC ACA C-3′. β-actin and Ppp1a were used as housekeeping genes for PCR: β-actin forward, 5′-CTA AGG CCA ACC GTG AAA AG-3′ and β-actin reverse, 5′-ACC AGA GGC ATA CAG GGA CA-3′, Ppp1a forward, 5′-GTG GTC TTT GGG AAG GTG AA-3′ and Ppp1a reverse, 5′-TTA CAG GAC ATT GCG AGC AG-3′.
Detection of enzymatic DDO activity
The tissues were homogenized in five volumes of cold 10 mM sodium pyrophosphate buffer (pH 8.3) containing protease inhibitors (Nacalai Tesque, Kyoto, Japan) using a BioMasher II (Nippi Inc., Tokyo, Japan). The homogenates were dialyzed for 1 day at 4 °C against 1 L of 10 mM sodium pyrophosphate buffer (pH 8.3) containing 2 mM EDTA, 5 mM 2-mercaptoethanol, and 10% (v/v) glycerol. The buffer was changed once during dialysis. The dialyzed homogenates were recovered and centrifuged at 20,000×g for 10 min at 4 °C to pellet any denatured proteins during the dialysis. The supernatants were recovered and stored at − 80 °C until use.
The protein concentration of dialyzed homogenates was determined using a Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA) with bovine serum albumin as a standard. DDO activity was determined using a colorimetric assay that detected production of 2-oxo acids, as previously described (Yamada et al. 1988) with some modifications. Appropriate amounts (60–300 μg) of the dialyzed homogenates were added to a reaction mixture containing 40 mM sodium pyrophosphate buffer (pH 8.3), 60 μM FAD, 33 ng/μL catalase from Aspergillus niger (Sigma-Aldrich), and 50 mM d-Asp in a final volume of 50 μL, and incubated at 37 °C. The incubation times with P90 whole brain homogenates and the others were 30 and 60 min, respectively. Subsequently, 4 μL of 100% (w/v) trichloroacetic acid (TCA) was added to stop the reaction. After centrifugation at 20,000×g for 10 min, a 50 μL portion of the supernatant was reacted with 33 μL of 0.1% (w/v) 2,4-dinitrophenylhydrazine in 2 M HCl at 37 °C for 15 min. A 250 μL of 3.75 M NaOH was added to the solution, following by centrifugation at 20,000×g for 10 min and standing for 10 min. The 2-oxo acid (oxaloacetic acid) products were quantified by measuring the absorbance of a 200 μL portion of the supernatant at 445 nm against a blank mixture lacking d-Asp.
High-performance liquid chromatography analysis
Amino acid levels in mouse brain tissue samples were analyzed using HPLC as previously reported (Nuzzo et al. 2017, 2019a), with minor modifications. Briefly, samples were homogenized in 1:10 (w/v) 0.2 M TCA, sonicated (3 cycles, 10 s each) and centrifuged at 13,000×g for 20 min. The precipitated protein pellets were stored at − 80 °C for protein quantification, while the supernatants were neutralized with NaOH and subjected to precolumn derivatization with o-phthaldialdehyde/N-acetyl-l-cysteine. Diastereoisomer derivatives were resolved on a Simmetry C8 5-μm reversed-phase column (Waters, 4.6 × 250 mm). Identification and quantification of d-Ser, l-Ser, d-Asp, l-Asp, and l-Glu were based on retention times and peak areas, compared with those associated with external standards. The identity of peaks was confirmed by adding known amounts of external standards. The identity of d-Asp peak was also evaluated by the selective degradation catalyzed by a recombinant human DDO (hDDO) (Katane et al. 2017, 2018). The hDDO enzyme (12.5 μg) was added to the samples, incubated at 30 °C for 3 h, and subsequently derivatized. Total protein content of homogenates was determined using the Bradford assay method, after resolubilizing the TCA-precipitated protein pellets. The total concentration of amino acids detected in tissue homogenates was normalized by the total protein content and expressed as nmol/mg protein.
Western blotting
Preparation and immunoblotting were performed as previously described (Nuzzo et al. 2019b). Frozen, powdered samples from post-mortem brains were sonicated in 1% SDS and boiled for 10 min. Proteins were separated by SDS-PAGE and electroblotted onto PVDF membranes (GE-Healthcare). Immunodetections were accomplished using the following antibodies: anti-GluN1 (1:1000, Cell Signaling Technology, Beverly, MA, USA), anti-GluN2A (1:1000, Sigma, St. Louis, MO, USA), anti-GluN2B (1:500, Cell Signaling Technology), anti-mGluR5 (1:1000, Abcam, Cambridge, UK), anti-GluA1 (1:500, Merck Millipore, Darmstadt, Germany), anti-GluA2/3 (1:500, Merck Millipore), anti-PSD-95 (1:1000, BD Biosciences, Franklin Lakes, NJ, USA), anti-Homer1b/c (1:1000, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-α-tubulin (1:50,000, Sigma). Blots were then incubated in horseradish peroxidase-conjugated secondary antibodies. Immunoreactivity was detected by enhanced chemiluminescence (ECL) (GE-Healthcare) and quantified by Quantity One software (Bio-Rad). Optical density values were normalized to α-tubulin for variations in loading and transfer.
Histochemistry staining of mouse brain tissues
Brains from R26+/+, R26Ddo/+ and R26Ddo/Ddo mice were cut in serial sagittal sections (25 µm-thick) at a freezing microtome, mounted on electrostatic glass slides and let dry. After a rehydration through a series of descending ethyl alcohols (100%, 70%, and 50%) and distilled water, sections were stained for 3 min, at RT, in a 0.5% solution of cresyl violet. After a rinse in distilled water, sections were dehydrated with a series of ascending ethyl alcohol (50%, 70%, and 100%) and xylene, let dry and coverslipped with Eukitt mounting medium.
Primary antibodies
The following antibodies were used for immunohistochemistry: mouse anti-NeuN, clone 60 (Millipore, Temecula, CA, USA), diluted 1:100; rat anti-myelin basic protein (MBP) (Millipore), diluted 1:250; rabbit anti-somatostatin (SST) (Sigma-Aldrich, Saint Lous, MO, USA), diluted 1:90; mouse anti-parvalbumin (PV) (Sigma-Aldrich), diluted 1:2000; rabbit anti-calbindin 28 K, diluted 1:12,000.
Immunohistochemistry staining of mouse brain tissues
Brains from R26+/+, R26Ddo/+ and R26Ddo/Ddo mice were cut in serial coronal sections (25 μm-thick) at a freezing microtome and treated as free-floating tissues. Cerebella of the same mice were cut apart, according to a sagittal plane. After blocking endogenous peroxidases (10 min at RT) in 0.3% H2O2, 10% methanol (in 0.5 M Tris/HCl, pH 7.6), sections were rinsed in buffer, incubated (1 h, at RT) in 5% defatted dry milk (DM), 0.5% Triton X-100 in 0.5 M Tris/HCl, pH 7.6, to block non-specific binding sites, and then incubated (48 h at 4 °C) in one of the primary antibodies diluted in 1% DM, 0.2% Triton X-100 in 0.5 M Tris/HCl, pH 7.6. Antigen–antibody binding sites were revealed by the peroxidase (HRP) immuno-enzymatic method. Briefly, after a thorough rinse in 0.5 M Tris/HCl, sections were incubated (1 h, at RT) with either a Goat anti-rabbit IgG (diluted 1:100) or Goat anti-mouse IgG (diluted 1:100) secondary antibody (Jackson Immunoresearch Europe Ltd, Cambridgeshire, UK), depending on the host of the primary antibody. For the rat primary antibody, we used a Goat anti-rat IgG-HRP conjugated secondary antibody (Jackson Immunoresearch), diluted 1:200, for 2 h, at RT. After a rinse in Tris/HCl, sections incubated with the first two secondary antibodies were further incubated (1 h, at RT) with either a Rabbit IgG-PAP (peroxidase-anti-peroxidase complex) or a Mouse IgG-PAP (Jackson Immunoresearch). After a final rinse in 0.5 M Tris/HCl, antibody binding sites for all three conditions were revealed by incubating sections (20 min at RT) in 0.05% 3,3′-diaminobenzidine (DAB) and 0.03% H2O2 in 0.5 M Tris/HCl, pH 7.6. Negative controls were obtained by omitting the primary antibodies and were always immunonegative. Sections were transferred in a 0.1% Cresyl gel solution (0.1% gelatin, 40% ethanol in dH2O) just before being mounted on glass slides, air-dried and permanently coverslipped with Eukitt mounting medium.
Microscopic image acquisition and quantitative image analysis
The number of SST- and PV-positive neurons was counted at the medial PFC level using the Image J software. Serial sections were arbitrarily subdivided into three groups, based on their distance from Bregma: anterior (2.96–1.70 mm from Bregma), medial (1.54–0.14 mm from Bregma) and posterior (0.02 to − 0.82 mm from Bregma), according to The Mouse Brain in Stereotaxic Coordinates (Paxinos and Franklin 2001). For each section, random areas were traced at the medial PFC level and the number of immunopositive cells contained in each area was counted. Final data were expressed as the mean number of cells/106 µm2 ± SEM. Counts were performed genotype blindly.
Morris water maze
The hidden-platform version of the Morris water maze test was conducted in a circular pool (100 cm in diameter) with a platform (8 cm in diameter) submerged 1 cm beneath the water surface (at 21 ± 1 °C). The hidden-platform version was performed according to a previous protocol (Errico et al. 2008b). The pool was surrounded by three-dimensional visual cues and contained opaque water. Mice were gently handled 5 min per day for a week before the experiment. The acquisition phase consisted of two sessions per day (3 h interval between sessions) over a 5-day period. Each session was composed of four trials with an inter-trial interval of approximately 5 min. On day 6, the platform was moved to the opposite position and the reversal learning was monitored for five additional days. Four probe tests were performed (two tasks per each training phase) in which animals were allowed to swim for 60 s in the absence of the platform, to evaluate time-dependent memory retention of mice. During acquisition phase, the first test (probe 1) was performed before starting the 4th day of training, while the second (probe 2) was conducted at day 6, before the first session of the reversal phase. During reversal phase, the first retention task (probe 3) was done before starting the 9th day of training, while the second (probe 4) was conducted at the end of the reversal phase, at day 11. Each retention test was conducted about 18 h after the second session of the previous day. The escape latency in the acquisition and reversal phase and the duration of time spent in each quadrant in the probe tests were recorded through a computerized video tracking system (Videotrack, ViewpointS.A., Champagne-au-Mont-d’Or, France).
Novel object recognition
The experimental procedure was in accord to a previous protocol (Errico et al. 2011b). Mice were gently handled 5 min per day for a week before the experiment.To provide mice the familiarity to the testing environment, a 3-day habituation phase was conducted by exposing each animal to the experimental cage (40 cm × 40 cm Plexiglas chamber with white walls and black floor) for at least 20 min/day. On the training session (day 4), the box was enriched with two identical green, squared Lego objects (3 cm × 6 cm × 6 cm), positioned in the back left and right corners of the apparatus. Each animal was placed in the middle point of the wall opposite to the sample objects and left to freely explore and familiarize with the objects for 15 min. Twenty-four hours later (testing session), one of the two objects (familial) was substituted by a new one (novel), different in color and shape (a grey, metal pyramid, 5 cm × 5 cm × 6 cm). Each mouse was placed in the apparatus and left free to explore it for 10 min. Both training and testing phases were video recorded and the time spent exploring each of the two objects was measured by a genotype-blind experimenter. Sniffing and touching the object were considered to be exploratory behavior. During behavioral sessions, objects were fixed with tape to the floor so that the animals could not move them and none of the objects used evoked innate preference. The exploration time (%) that animals spent interacting with the novel object over the total exploration time (e.g. [novel/(familial + novel)] × 100) during the testing phase was measured. Mice that had less than 8 s of exploration during the training phase were excluded from the analysis.
Statistical analyses
Statistical analyses were performed in GraphPad Prism 7. All data were expressed as mean ± standard error of the mean (SEM). Sample size of each experiment is indicated in the respective figure legend. Statistical assessment of gene expression, enzymatic activity, and amino acids content were performed both during ontogenesis and at adulthood. Statistical analyses during ontogenesis were performed by two-way ANOVA (genotype × age) with age as repeated measure, followed by Fisher’s post hoc analysis between genotypes, when required. Data on single adult brain regions were analyzed by one-way ANOVA (genotype effect), followed by Fisher’s post hoc analysis, when required. Statistical analysis of western blotting was performed by Student’s t test. Statistical analysis of PV- or SST-positive interneurons was performed by one-way ANOVA (genotype effect), followed by Fisher’s post hoc analysis, when required. Learning performance during both acquisition and reversal phase in the hidden platform version of the Morris water maze was analyzed by two-way ANOVA (genotype × days) with days as repeated measure. Memory abilities in the probe tests were analyzed by one-way ANOVA with repeated measures (quadrant effect), followed by Fisher’s post hoc analysis between quadrants, when required. Finally, recognition memory in the object recognition test was analyzed by two-way ANOVA (genotype × object), followed by Fisher’s post hoc comparisons between objects or genotypes, when required. Asterisks denote statistical significance as calculated by the specified statistical tests (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Results
Generation of knockin mice with embryonic Ddo expression
A fundamental hallmark of d-Asp lies within its transient prenatal occurrence in the mammalian brain (Dunlop et al. 1986; Hashimoto et al. 1993, 1995; Neidle and Dunlop 1990; Sakai et al. 1998; Wolosker et al. 2000; Punzo et al. 2016). To understand the biological significance of this molecule in the developing brain, we generated a novel Ddo knockin mouse model enabling the precocious expression of DDO enzyme and, in turn, the depletion of embryonic d-Asp storage. To achieve this goal, we targeted an inducible Ddo-LacZ cassette (LoxP-Stop-LoxP-Ddo-IRES-LacZ-polyA: LSL-DdoLacZ) into the genomic locus of Rosa26 (Fig. 1a). In this way, the removal of the triple poly-A sequence (stop cassette) by Cre/loxP recombination system is expected to enable the ectopic expression of Ddo under the regulatory control of the constitutive Rosa26 promoter. The targeting vector was electroporated in the E14Tg4a2 embryonic stem cell line and 100 G418-resistant colonies were analyzed by PCR and Southern blotting to ascertain the correct homologous recombination (Fig. 1b, c). To generate mice carrying R26LSL−DdoLacZ allele, one transformed clone was injected into B6 blastocysts after removal of the neomycin-resistance cassette. These animals were then crossed with Hprt/Cre mice to remove the stop cassette and allow the constitutive expression of the Ddo exogenous gene (Fig. 1a). The resulting knockin mice (R26Ddo/+) were interbred to generate homozygous animals (R26Ddo/Ddo) and genotyped by PCR (Fig. 1d). Both heterozygous and homozygous knockin mice were viable, fertile, and undistinguishable from control littermates (Fig. 1e).
Dose-dependent expression of Ddo mRNA in the brain of Ddo knockin mice
To verify that extra copies of Ddo were actually reflected into increased transcription, we detected Ddo mRNA levels by quantitative real time PCR (qRT-PCR) in the whole developing brain and in different adult cerebral regions of the mutant mice.
First, we examined Ddo transcript levels in the whole brain of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice during ontogenesis, in a time window ranging from E16.5 to P90 stage (Fig. 2a). In accordance with the presence of additional copies of Ddo in knockin mice, we observed a significant dose-dependent increase in Ddo mRNA levels [two-way ANOVA, F(2,35) = 186.00, p < 0.0001]. Further analysis indicated that Ddo expression was significantly upregulated in both R26Ddo/+ and R26Ddo/Ddo mice, compared to R26+/+ littermates, at different time points (R26Ddo/+: p < 0.0001 at E16.5, E18.5, P3, and P90; p < 0.01 at P7 and P14; R26Ddo/Ddo: p < 0.0001 at each age; Fisher’s post hoc comparison). Two-way ANOVA also showed a significant effect of age [F(3,35) = 3.36, p = 0.0139] and age × genotype interaction [F(10,35) = 2.25, p = 0.0375], as a consequence of a different age-dependent Ddo transcription profile among genotypes. Indeed, physiological expression of Ddo gene increased over time in the whole brain of R26+/+ mice [one-way ANOVA, F(5,12) = 19.08, p < 0.0001], as previously demonstrated in Ddo+/+ mice (Punzo et al. 2016). In contrast, Ddo mRNA levels in R26Ddo/Ddo animals were steadily upregulated [F(5,12) = 2.03, p = 0.1466], as a result of the stable cerebral Ddo transcription driven by the Rosa26 promoter. Then, we focused on Ddo mRNA expression in different brain regions of adult (P90) knockin mice, including PFC, hippocampus, striatum, and cerebellum (Fig. 2b–e). In each brain area, we found a significant effect of genotype [one-way ANOVA: PFC, F(2,10) = 14.35, p = 0.0012; hippocampus, F(2,10) = 25.53, p = 0.0001; striatum, F(2,10) = 23.26, p = 0.0002; cerebellum, F(2,10) = 12.89, p = 0.0017], as Ddo mRNA content was increased according to the allelic Ddo copy number.
Analysis of Ddo, Daao, and Sr mRNA content in the brain of Ddo knockin mice. mRNA expression of aDdo, fDaao, and kSr in the whole brain of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (n = 3/genotype) at embryonic day 16.5 (E16.5), E18.5, and postnatal day 3 (P3), P7, P14 and P90. mRNA levels of b–e Ddo, g–jDaao and l–oSr in the b, g, l prefrontal cortex, c, h, m hippocampus, d, i, n striatum and e, j, o cerebellum of R26+/+ (n = 3), R26Ddo/+ (n = 5) and R26Ddo/Ddo mice (n = 5) at P90. Ddo, Daao and Sr mRNA levels were detected by quantitative RT-PCR, normalized to the mean of two housekeeping genes (β-actin and Ppp1a) and expressed as 2−ΔCt. N.D. indicates that Daao transcript was not detectable. *p < 0.05, **p < 0.01, ***p < 0.0001, compared to R26+/+ mice (Fisher’s post hoc comparisons). #p < 0.05, ##p < 0.01, compared to R26Ddo/+ mice (Fisher’s post hoc comparisons)
d-amino acid oxidase and serine racemase gene expression in Ddo knockin brains
Previous studies indicated that d-Asp and d-Ser metabolism have partially overlapping pathways, as serine racemase (SR) enzyme, which is responsible for d-Ser biosynthesis (Wolosker et al. 1999), has been demonstrated to take part also in d-Asp production (Horio et al. 2013; Ito et al. 2016). In light of this, we evaluated whether increased Ddo transcription in knockin brains affected the expression of the genes regulating d-Ser metabolism, such as d-amino acid oxidase (Daao) (Pollegioni et al. 2007) and Sr. However, we did not observe any genotype effect in mRNA levels of these genes [Daao: F(2,35) = 0.14, p = 0.8652; Sr: F(2,35) = 2.51, p = 0.0958], suggesting that their ontogenetic regulation is not influenced by Ddo constitutive expression. On the other hand, in line with previous studies in mouse and human brains (Jagannath et al. 2017; Wang and Zhu 2003), we found a significant increase in the expression of both Daao and Sr genes over time [Daao: F(5,35) = 69.73, p < 0.0001; Sr: F(5,35) = 132.70, p < 0.0001] (Fig. 2f, k). In particular, Daao mRNA levels were undetectable until the first postnatal week, before considerable increase afterwards. Interestingly, we confirmed that Daao gene expression in the adult brain is confined to the cerebellum (Jagannath et al. 2017; Wang and Zhu 2003; Cuomo et al. 2019) (Fig. 2g–j), where no significant changes were observed among different genotypes [one-way ANOVA, F(2,10) = 0.28, p = 0.7593]. On the other hand, Sr gene was expressed in all of the brain regions tested (Cuomo et al. 2019), with no difference among different genotypes (Fig. 2l–o).
Enzymatic DDO activity in the brain of Ddo knockin mice
Furthermore, we assessed whether upregulated Ddo mRNA levels in knockin mice resulted in higher enzymatic DDO activity in the brain of these animals. In accordance with the increased Ddo gene expression in Ddo knockin mice, we observed that the enzymatic activity of DDO in the whole brain of these animals is significantly increased in a dose-dependent manner [two-way ANOVA, F(2,24) = 175.80, p < 0.0001] (Fig. 3a). Moreover, we also found a significant age effect [F(3,24) = 0.14, p < 0.0001] and genotype × age interaction [F(6,24) = 14.65, p < 0.0001] on the DDO enzymatic activity in mutant mice. Interestingly, we observed a substantial decrease in the enzymatic DDO activity of R26Ddo/+ and R26Ddo/Ddo mice during adulthood [one way ANOVA: R26Ddo/+, F(3,8) = 0.14, p = 0.0004; R26Ddo/Ddo, F(3,8) = 23.19, p = 0.0003]. This was in contrast to age-dependent increase of enzymatic DDO activity observed in R26+/+ mice [F(3,8) = 5.57, p = 0.0233] and in rats (Van Veldhoven et al. 1991). Overall, we found a greater enzymatic activity in the whole brain of Ddo knockin mice at all time points, comparing to their R26+/+ littermates (Fig. 3a). However, while at P3, we found about 100 times higher DDO activity in R26Ddo/Ddo vs R26+/+ mice (5.260 vs 0.053 nmol/min/mg protein), at P90, we only observed around sevenfolds of such an increase (1.073 vs 0.163 nmol/min/mg protein). Similar to the whole brain, different adult brain regions also showed a significantly higher DDO activity in R26Ddo/+ and R26Ddo/Ddo mice, compared to R26+/+ littermates [one-way ANOVA: prefrontal cortex, F(2,9) = 20.31, p = 0.0005; hippocampus, F(2,9) = 20.85, p = 0.0004; striatum, F(2,9) = 122.60, p < 0.0001; cerebellum, F(2,9) = 13.07, p = 0.0022] (Fig. 3b–e).
Detection of enzymatic DDO activity in the brain of Ddo knockin mice. Enzymatic activity of DDO a in the whole brain of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (n = 3/genotype) at embryonic day 16.5 (E16.5), E18.5, and postnatal day 3 (P3), P7, P14, and P90. DDO activity in the b prefrontal cortex, c hippocampus, d striatum, and e cerebellum of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (n = 4/genotype) at P90. Values are expressed as the mean ± SEM. **p < 0.01, ***p < 0.0001, compared to R26+/+ mice (Fisher’s post hoc comparisons). #p < 0.05, ##p < 0.01, compared to R26Ddo/+ mice (Fisher’s post hoc comparisons)
Cerebral d-aspartate content in Ddo knockin mice
We assessed whether increased cerebral DDO activity in R26Ddo/+ and R26Ddo/Ddo brains affected the endogenous levels of its substrate, d-Asp. We first performed HPLC detection of d-Asp in the whole brain of R26+/+, R26Ddo/+ and R26Ddo/Ddo animals during ontogenesis, from E16.5 to P90 (Fig. 4a). We observed a significant effect of genotype [two-way ANOVA, F(2,32) = 112.20, p < 0.0001] and, additionally, a significant age effect and genotype × age interaction [age: F(5,32) = 9.58, p < 0.0001; age × genotype: F(10,32) = 8.89, p < 0.0001]. In line with previous data in rodents and humans (Hashimoto et al. 1993; Punzo et al. 2016; Sakai et al. 1998; Wolosker et al. 2000), d-Asp levels physiologically decreased in the whole brain of R26+/+ mice [one-way ANOVA, F(5,12) = 10.41, p = 0.0005]. Conversely, the increase in cerebral DDO activity in R26Ddo/+ and R26Ddo/Ddo animals produced a strong reduction of this d-amino acid content, which remains constant over time [R26Ddo/+: F(5,11) = 1.73, p = 0.2080; R26Ddo/Ddo: F(5,9) = 1.54, p = 0.2709]. Remarkably, at all ages tested, d-Asp reduction was always greater than 90%, and independent from the allelic Ddo dosage. In agreement with the results obtained in the whole brain, we also observed a significant decrease in d-Asp levels in all adult brain regions of both R26Ddo/+ and R26Ddo/Ddo mice, compared to their control littermates [one-way ANOVA: PFC, F(2,6) = 23.04, p = 0.0015; hippocampus, F(2,6) = 32.32, p = 0.0006; striatum, F(2,6) = 4.70, p = 0.0591; cerebellum, F(2,6) = 13.75, p = 0.0057] (Fig. 4b–e). Although we found a significant change between R26Ddo/+ and R26Ddo/Ddo mice only in the hippocampus (Fig. 4c), a reduction trend of d-Asp content in R26Ddo/Ddo animals was apparent in each adult brain area (Fig. 4b–e).
Detection of free d-Asp and l-Asp content in the brain of Ddo knockin mice. Analysis of ad-Asp and fl-Asp levels, as well as kd-/total Asp ratio in the whole brain of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (n = 3/genotype) at embryonic day 16.5 (E16.5), E18.5, postnatal day 3 (P3), P7, P14 and P90. Analysis of b–ed-Asp and g–jl-Asp levels, as well as l–od-/total Asp ratio in the b, g, l prefrontal cortex, c, h, m hippocampus, d, i, n striatum and e, j, o cerebellum of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (n = 3/genotype) at P90. *p < 0.05, **p < 0.01, ***p < 0.0001, compared to R26+/+ mice (Fisher’s post hoc comparisons). #p < 0.05, ##p < 0.01, compared to R26Ddo/+ mice (Fisher’s post hoc comparisons). Values are expressed as the mean ± SEM. All the amino acids were detected in a single run by HPLC and expressed as nmol/mg protein, while the ratio is expressed as percentage (%)
Then, we measured the levels of l-Asp, both in the whole brain during ontogenesis and within adult brain regions of R26+/+, R26Ddo/+ and R26Ddo/Ddo mice. We did not find any genotype-dependent variation of this l-enantiomer in the whole brain during ontogenesis [two-way ANOVA, genotype: F(2,35) = 0.56, p = 0.5786; genotype × age: F(10,35) = 0.39, p = 0.9421] (Fig. 4f). Similarly, no genotype effect was observed in any adult brain regions (one-way ANOVA, p > 0.05 in each brain region) (Fig. 4g–j). Altogether, these results suggested that deregulated Ddo gene expression through knockin approach selectively perturbs d-Asp but not l-Asp catabolism.
Moreover, in line with a selective change in cerebral d-Asp levels, we revealed significant effect of genotype [two-way ANOVA, F(2,36) = 392.90, p < 0.0001], age [F(5,36) = 73.62, p < 0.0001] and genotype × age interaction [F(10,36) = 62.08, p < 0.0001] in the ratio between d-Asp and total Asp (d-Asp + l-Asp) in the whole brain of mice during ontogenesis (Fig. 4k). Interestingly, in R26+/+ mice, the peak of relative d-Asp occurrence was found at E18.5 (p = 0.0137, compared to E16.5; p < 0.0001, compared to all other time points; Fisher’s post hoc comparison), when the d-enantiomer represented almost the 12% of total Asp amount. As in the whole brain, also in the adult brain regions analyzed, we found significantly different d-Asp /total Asp ratio among the different genotypes [one-way ANOVA: PFC, F(2,6) = 29.47, p = 0.0008; hippocampus, F(2,6) = 93.76, p < 0.0001; striatum, F(2,6) = 5.19, p = 0.0492; cerebellum, F(2,5) = 14.10, p = 0.0088] (Fig. 4l–o).
d-Serine, l-serine, and l-glutamate levels in the brain of Ddo knockin mice
We assessed whether the overall changes in d-Asp metabolism could affect the content of the other neuroactive d-amino acid, d-Ser, and of its precursor, l-Ser (Wolosker et al. 1999) in the brain of R26Ddo/+ and R26Ddo/Ddo mice. In line with unaltered Daao and Sr gene expression in mutants, statistical analysis of d-Ser content showed no genotype effect in the whole brain during ontogenesis [two-way ANOVA, F(2,35) = 0.08, p = 0.9202]. Conversely, we found a significant age-dependent increase in d-Ser levels [F(5,35) = 121.70, p < 0.0001] (Fig. 5a), which is in agreement with a previous evidence obtained in mouse cortex and striatum (Wang and Zhu 2003). d-Ser levels were also unaltered among different genotypes in all the adult brain regions (Fig. 5b–e). Similarly, no genotype-dependent changes in l-Ser levels were detected both in the whole brain during ontogenesis [F(2,35) = 0.54, p = 0.5877] (Fig. 5f) and in all investigated adult cerebral areas (Fig. 5g–j). Accordingly, also d-Ser/total Ser ratio was unaffected by the genotype in the whole brain [F(2,35) = 0.20, p = 0.8207] (Fig. 5k), as well as in different adult brain regions (p > 0.05) (Fig. 5l–o).
Detection of free d-Ser, l-Ser, and l-Glu content in the brain of Ddo knockin mice. Analysis of ad-Ser and fl-Ser levels, kd-/total Ser ratio, and pl-Glu levels in the whole brain of R26+/+, R26Ddo/+, and R26Ddo/Ddo mice (n = 3/genotype) at embryonic day 16.5 (E16.5), E18.5, postnatal day 3 (P3), P7, P14, and P90 (n = 3/genotype). Analysis of b–ed-Ser, g–jl-Ser levels, l–od-/total Asp ratio and q–tl-Glu levels in the b, g, l, q prefrontal cortex, c, h, m, r hippocampus, d, i, n, s striatum and e, j, o, t cerebellum of R26+/+, R26Ddo/+,and R26Ddo/Ddo mice (n = 3/genotype) at P90. Values are expressed as the mean ± SEM. All the amino acids were detected in a single run by HPLC and expressed as nmol/mg protein, while the ratio is expressed as percentage (%)
Finally, we measured the levels of the main excitatory amino acid, l-Glu, in the brain of knockin mice. We did not find a genotype effect during lifespan [F(2,35) = 1.06, p = 0.3583], although l-Glu levels changed in an age-dependent manner [F(5,35) = 11.96, p < 0.0001] (Fig. 5p). Likewise, no variations in l-Glu content occurred among genotypes, in all investigated brain areas (p > 0.05) (Fig. 5q–t).
Altogether, these results highlighted that constitutive d-Asp depletion in Ddo knockin mice does not affect the metabolism of the main neuroactive molecules involved in NMDAR-dependent transmission, d-Ser and l-Glu.
Glutamatergic NMDA, AMPA, and mGluR5 receptors and post-synaptic density proteins levels in the adult Ddo knockin brain
We analyzed whether early d-Asp depletion may affect in mutant brains the levels of the NMDAR subunits, (GluN1, GluN2A, and GluN2B), AMPAR subunits (GluA1 and GluA2/3), and mGluR5 receptors, which are known to be affected by this d-amino acid (Fagg and Matus 1984; Olverman et al. 1988; Errico et al. 2018; Gong et al. 2005; Molinaro et al. 2010). This analysis was performed by western blotting in different brain regions of adult R26Ddo/Ddo mice (3/4-month-old), such as the PFC (Fig. 6a–f), hippocampus (Fig. 6g–l), striatum (Fig. 6m–r), and cerebellum (Fig. 6s–x). Interestingly, statistical analysis indicated that the expression levels of each of these proteins were undistinguishable between R26Ddo/Ddo and R26+/+ control animals, in each brain area analyzed.
Expression levels of glutamatergic receptors in the brain of Ddo knockin mice. The protein expression levels of the NMDAR subunits a, g, m, s GluN1, b, h, n, t GluN2A and c, i, o, u GluN2B, AMPAR subunits d, j, p, v GluA1 and e, k, q, w GluA2/3, and f, l, r, x metabotropic glutamate receptor, mGluR5, were detected in the a–f prefrontal cortex, g–l hippocampus, m–r striatum and s–x cerebellum of control R26+/+ and R26Ddo/Ddo mice by western blotting. Tubulin was used to normalize for variations in loading and transfer. The numerosity of samples examined was as follows: prefrontal cortex and hippocampus, n = 7 mice/genotype for each immunodetection; striatum, n = 7 mice/genotype for each immunodetection except GluN2A (n = 7 R26+/+, n = 6 R26Ddo/Ddo) and mGluR5 (n = 7 R26+/+, n = 5 R26Ddo/Ddo); cerebellum, n = 6 R26+/+, n = 7 R26Ddo/Ddo for GluN1; n = 7 mice/genotype for GluN2A and GluA1; n = 6 mice/genotype for GluN2B, n = 6 R26+/+, n = 7 R26Ddo/Ddo for GluA2/3, n = 5 mice/genotype for mGluR5
To further investigate the influence of early d-Asp depletion on glutamatergic synapse composition, we also tested in the same brain samples the levels of the post-synaptic density proteins, PSD-95, and Homer 1b/c (Fig. 7a–h), known to be critically involved in the regulation of NMDA and mGluR5 receptors signaling (de Bartolomeis and Tomasetti 2012). Overall, western blot analysis showed no main genotype effect for both glutamatergic scaffolding proteins in each brain region tested. Taken together, our data exclude the existence of gross alterations in glutamatergic synapse composition in adult R26Ddo/Ddo mutant brains, developed without a physiological content of d-Asp.
Expression levels of PSD-95 and Homer 1b/c in the brain of Ddo knockin mice. The expression levels of the post-synaptic density proteins a, c, e, g PSD-95 and b, d, f, h Homer 1b/c were detected in the a, b prefrontal cortex, c, d hippocampus, e, f striatum and g, h cerebellum of control R26+/+ and R26Ddo/Ddo mice by western blotting. The numerosity of samples examined was as follows: n = 8 mice/genotype for each immunodetection and in each brain region except the prefrontal cortex (n = 7 R26+/+, n = 8 R26Ddo/Ddo for each immunodetection)
Effect of early d-aspartate deprivation on adult brain morphology
We evaluated whether early depletion of cerebral d-Asp levels could affect adult brain morphology and the overall neuronal organization and distribution in adult brain regions of Ddo knockin mice (3/4-month-old). A first analysis on sagittal brain cryosections stained with cresyl violet did not reveal any gross anatomical difference (i.e. brain size and structural organization) among R26+/+, R26Ddo/+ and R26Ddo/Ddo mice (Fig. 8a–c).
Histological, NeuN, and calbindin immunohistochemical analyses on brain sections of Ddo knockin mice. a–c Representative sagittal brain sections, taken approximately 1.80–2.04 mm lateral to the midline, according to The Mouse Brain Atlas in Stereotaxic Coordinates (Paxinos and Franklin 2001), stained with cresyl violet of aR26+/+, bR26Ddo/+ and cR26Ddo/Ddo mice (n = 3/genotype), taken approximately 1.80–2.04 mm lateral to the midline, according to The Mouse Brain Atlas in Stereotaxic Coordinates (Paxinos and Franklin 2001). Main brain regions are indicated: CPu caudate putamen, Crb cerebellum, Ctx cortex, Hip hippocampus, OB olfactory bulb. Scale bar 500 μm. d–f, h–j, l–n, p–r NeuN immunostaining on d–f, h–j, l–n brain coronal and p–r cerebellar sagittal serial sections [s representative sections 0.60–0.84 mm lateral to the midline] of (d, h, l, pR26+/+, e, i, m, q) R26Ddo/+ and f, j, n, rR26Ddo/Ddo mice (n = 3/genotype). Brain coronal sections analyzed were obtained from the d–f medial prefrontal cortex (mPFC) [g representative section 0.98–0.86 mm from Bregma], h–j striatum [k representative section 0.98–0.86 mm from Bregma], and l–n hippocampus [o representative sections − 1.94 mm to − 2.06 from Bregma]. t–y Calbindin immunolabeling on cerebellar sagittal sections preferentially labeling Purkinje cell somata and their dendrites in the molecular layer (w–y, higher magnification), as well as fibers running through the cerebellar nuclei (MedDL, IntP). Scale bars: mPFC, striatum, hippocampus: 200 μm; cerebellum: 500 μm; Purkinje cell higher magnifications: 50 μm II, III, IV/V, VI, VII, VIII, IX, and X cerebellar lobules, CA1, CA2, and CA3 cornu ammonis areas 1, 2 and 3, Cg1 and Cg2 cingulate cortex, areas 1 and 2, CPu caudate putamen (striatum), DG dentate gyrus, ec external capsule, M1 and M2 primary and secondary motor cortices, MEdDL medial cerebellar nucleus, ML molecular layer, IntP interposed cerebellar nucleus, PC Purkinje cells
We successively performed NeuN immunolabeling on brain serial coronal sections to acquire information on the general neuronal organization and distribution in Ddo knockin mice. We observed no differences among genotypes in cell density and distribution in either of the four analyzed main brain regions, such as the medial PFC (Fig. 8d–f), striatum (Fig. 8h–j), hippocampus (Fig. 8l–n), and cerebellum (Fig. 8p–r). In addition, immunolabeling for calbindin, a major marker of cerebellar Purkinje cells specifically chosen to investigate whether Ddo knockin mice could undergo a disorganization of the cerebellar layering, revealed no significant differences among the three genotypes (Fig. 8t–y), including the development of Purkinje cell dendritic arborization in the molecular layer (Fig. 8w–y).
Based on a previous report indicating a role for d-Asp on myelination processes (de Rosa et al. 2019), we also evaluated the expression of the myelin basic protein (MBP) in adult Ddo knockin brains. Similar to NeuN, no differences in MBP immunolabeling were observed among the three genotypes, at any brain level (Fig. 9a–c).
Immunolabeling for myelin basic protein, parvalbumin, and somatostatin on medial prefrontal cortex sections of Ddo knockin mice. Brain coronal sections from a, e, iR26+/+ (n = 2), b, f, jR26Ddo/+ (n = 2) and c, g, kR26Ddo/Ddo mice (n = 3) were immunolabeled for a–c myelin basic protein (MBP), e–g parvalbumin and i–k somatostatin. d MBP-positive representative sections are taken 0.50 mmm to 0.26 mm from bregma, while those of parvalbumin and somatostatin immunolabeling, 2.96–1.70 mm from bregma. Quantitative analysis of h parvalbumin- and l somatostatin-positive interneurons. *p < 0.05, compared to R26+/+ mice (Fisher’s post hoc comparisons). Data are expressed as the mean ± SEM of immunopositive neurons/106 μm2. Scale bars MBP, 500 μm; parvalbumin and somatostatin, 200 μm; insets: 50 μm. aca anterior commissure, anterior, cg cingulum, Cg1 and Cg2 cingulate cortex, areas 1 and 2, CPu caudate putamen (striatum), ec external capsule, f fornix, LSD lateral septal nucleus, dorsal part, LSI lateral septal nucleus, intermediate, LV lateral ventricle, M1 and M2 primary and secondary motor cortices
In addition, we assessed the state of the inhibitory cortical pathways in the medial PFC of Ddo knockin mice. To this aim, we performed a quantitative analysis of parvalbumin (PV) and somatostatin (SST) immunostaining (Fig. 9e–l), two neuropeptides expressed by different subgroups of GABAergic interneurons (Tremblay et al. 2016). Interestingly, we observed significantly higher number of PV-positive interneurons in R26Ddo/+ and R26Ddo/Ddo mice, compared to R26+/+ littermates [one-way ANOVA, F(2, 5) = 6.41, p = 0.0417; p < 0.05, Fisher’s post hoc comparison) (Fig. 9h). Remarkably, the increased number of PV-positive interneurons in Ddo knockin mice showed a rostrocaudal gradient as it tends to decrease in the posterior regions of this brain area. Unlike PV immunolabeling, no differences among genotypes were observed in the number of SST-positive interneurons (Fig. 9l).
Depletion of d-aspartate during brain development is associated with increased memory in Ddo knockin mice at adulthood
Based on the influence of increased d-Asp levels on rodents’ memory (Errico et al. 2008b, 2011a, b; Topo et al. 2010), we investigated whether the substantial congenital d-Asp reduction could affect cognitive abilities of mutant mice at adulthood (3/4-month old). In this regard, we first evaluated spatial learning and memory of animals through a hidden-platform version of the Morris water maze (Fig. 10a). During the acquisition phase (days 1–5), we found comparable learning profiles among genotypes [two-way ANOVA, days, F(4,88) = 32.89, p < 0.0001, genotype, F(2,22) = 2.07, p = 0.1506]. To dissect the spatial memory of mice, we carried out two retention tests, after a short (3 days) (Fig. 10b) or long (5 days) (Fig. 10c) training period. Interestingly, after 3-day acquisition (probe 1), while R26+/+ and R26Ddo/+ animals showed a random quadrant search, R26Ddo/Ddo mice were already able to direct the platform search in the goal quadrant [one-way ANOVA, F(3,21) = 7.07, p = 0.0018; goal vs left, p < 0.05, goal vs right or opposite, p < 0.01, Fisher’s post hoc comparison] (Fig. 10b). Conversely, after a longer training exposure (5-day training, probe 2), all mice spent a significantly higher percentage of time in the target quadrant, independently from the genotype (Fig. 10c). Then, we moved the platform to the opposite position to analyze the ability of mice to erase the previous spatial information and plan a new navigation strategy. Also in the reversal phase (days 6–10), we revealed comparable learning abilities among genotypes (Fig. 10a). Indeed, statistical analysis showed a significant days effect [two-way ANOVA, F(4,88) = 15.59, p = 0.0008], but no differences between genotypes [F(2,22) = 0.2295, p = 0.7968]. During the first probe trial of the reversal phase (3-day training, probe 3), R26Ddo/Ddo mice showed an improved spatial memory [F(3,21) = 6.33, p = 0.0031; new goal vs right, left or opposite, p < 0.01], while R26+/+ and R26Ddo/+ mice showed no search preference for quadrants (Fig. 10d). After two additional training days (5-day training, probe 4), all mice manifested a preferential search for the target quadrant, regardless of genotype (Fig. 10e).
Evaluation of cognitive abilities in Ddo knockin mice.aR26+/+ (n = 9), R26Ddo/+ (n = 8), and R26Ddo/Ddo mice (n = 8) were trained in a submerged platform version of the Morris water maze during acquisition phase (days 1–5), followed by a reversal phase (days 6–10). Escape time, expressed in seconds (s), was used as dependent variable in the acquisition and reversal phases. At the top is shown the position of the platform in the circular pool during both the acquisition and reversal phase. b–e Four probe tests were performed (two tasks per each training phase) in the absence of the platform. Search quadrant, expressed as percentage (%) of time, was used as dependent variable in the probe tests. *p < 0.05, **p < 0.01, ***p < 0.0001, at least, compared to right, left or opposite quadrant (Fisher’s post hoc comparison). fR26+/+ (n = 7), R26Ddo/+ (n = 7), and R26Ddo/Ddo mice (n = 6) were tested for their recognition memory through the novel object recognition test. Exploratory preference, expressed as percentage (%) of time spent on the novel object [novel object time/ (familial object time + novel object time)] × 100), was used as dependent variable. *p < 0.05, **p < 0.01, compared to the familiar object (Fisher’s post hoc comparison). #p < 0.05, compared to R26+/+ or R26Ddo/+ mice (Fisher’s post hoc comparison). Values are expressed as the mean ± SEM
We also assessed recognition memory of knockin mice through the novel object recognition test (Fig. 10f). In the retention session, performed 24 h after the first exposure to the objects, two-way ANOVA revealed no difference among genotypes [F(2,17) = 0.92, p = 0.4172] but significant object discrimination [F(1,17) = 16.23, p = 0.0009] and genotype × object interaction [F(2,17) = 3.64, p = 0.0483], indicating a different exploratory propensity for the novel object among genotypes. Indeed, further statistical analyses highlighted that, although all genotypes displayed preference for the novel object, R26Ddo/Ddo mice spent a significantly longer time in the exploration of the novel object, compared to R26+/+ and R26Ddo/+ littermates [one-way ANOVA, F(2,17) = 3.66, p = 0.0475; R26Ddo/Ddo vs R26+/+, p = 0.0338; R26Ddo/Ddo vs R26Ddo/+, p = 0.0263, Fisher’s post hoc comparison].
Discussion
To address the still unknown role of precocious d-Asp occurrence on brain morphology and behavior at adulthood, we generated a novel knockin mouse model that overexpresses DDO during embryonic phase with the aim of depleting the cerebral d-Asp storage during brain development. Our results confirmed the success of the genetic strategy, as we found persistently high Ddo mRNA expression, accompanied by a robustly increased enzymatic DDO activity in Ddo knockin brains, compared to wild-type littermates. Consistent with these data, a dramatic decline of d-Asp levels (up to 95%) was reported in the brain of mutant animals, in both prenatal and postnatal phases. Remarkably, Ddo gene targeting has a selective influence on d-Asp but not on l-Asp content, since the levels of the l-enantiomer did not change both in R26Ddo/+ and R26Ddo/Ddo mutant mice. Likewise, upregulated Ddo gene expression did not produce neurochemical changes in the content of l-Glu, d-Ser, and l-Ser in brain homogenates of Ddo knockin mice. In this regard, the absence of variations in d-Ser content is in agreement with unaltered cerebral Daao and Sr gene expression in mutant mice. Overall, these findings demonstrate a selective influence of genetic Ddo manipulation for d-Asp metabolism, leaving unaffected the homeostasis of the other detected amino acids, l-Asp, d-Ser, l-Ser, and l-Glu. In line with unaltered levels of the main endogenous ligands for NMDARs, AMPARs and mGluR5, we found that d-Asp depletion does not affect the expression levels of the NMDAR subunits GluN1, GluN2A and 2B, AMPA receptors subunits, GluA1 and GluA2/3, and mGluR5 in any of the brain regions of R26Ddo/Ddo mice analyzed, compared with R26+/+ controls. Similarly, the levels of the postsynaptic scaffolding proteins, PSD-95 and Homer 1b/c, are also unaltered in R26Ddo/Ddo brains. Taken together, these results suggest that depletion of d-Asp since early developmental phases does not alter the overall composition of the mouse glutamatergic synapse.
As previously reported, transiently high d-Asp occurrence in the embryonic brain (Dunlop et al. 1986; Hashimoto et al. 1993, 1995; Neidle and Dunlop 1990; Sakai et al. 1998; Wolosker et al. 2000; Punzo et al. 2016) suggests a putative role for this d-amino acid in cellular events involved in neural development, such as neurogenesis, proliferation, migration, differentiation, apoptosis, and survival. Indeed, many of these fundamental processes are critically regulated by the glutamatergic NMDA and mGlu5 receptors (Ikonomidou 2009; Jansson and Akerman 2014; Di Giorgi-Gerevini et al. 2005), which are the pharmacological target of d-Asp signaling (Errico et al. 2018). Based on this knowledge, we expected that prenatal d-Asp depletion impacted on neuroanatomical and cognitive features of adult mutant mice by altering early glutamatergic transmission. Differently to our expectations, adult Ddo knockin mice show a gross normal brain morphology. Consistently, no main neuroanatomical alterations or changes in the overall neuronal and myelin distribution were found among genotypes. However, at a more detailed neuroanatomical investigation, we revealed a significant increase in the number of PV- but not SST-positive interneurons in the PFC of Ddo knockin mice, compared to wild-type littermates. The development of these connection neurons proceeds through a complex series of steps, including neurogenesis, migration to the cerebral cortex, morphological maturation, and programmed cell death, which covers both prenatal and postnatal phases and impacts on the final number of adult interneurons (Lim et al. 2018). Although we still ignore whether d-Asp is involved in one (or more) of these developmental events and what is its abundance in this class of interneurons and their precursor cells, our evidence suggests for the first time an inhibitory role for early d-Asp in the control of the number of PV-positive interneurons.
Interestingly, the ratio of d-Asp to total Asp in the embryonic mouse brain is much lower than the human one. Indeed, at the peak of its abundance at the end of gestational phase, we found that d-Asp reaches only the 12% of total Asp in wild-type mouse brains. In contrast, previous studies on human embryonic PFC demonstrated that d-Asp levels even exceed those of l-Asp in the first trimester of pregnancy, reaching around 65% of total Asp (Hashimoto et al. 1993). So, the higher relative amount of d-Asp in human embryonic brain might indicate that this d-amino acid has a selective importance in human neural developmental processes. Therefore, we argue that d-Asp may have a species-specific role for brain development. In this line of reasoning, previous fMRI evidence supports a role for DDO on the gray matter size of the human PFC. Indeed, we revealed that in healthy humans a single-nucleotide polymorphism associated with reduced prefrontal expression of DDO, predicts enhanced prefrontal gray matter volume, and greater prefrontal activity during working memory tasks (Errico et al. 2014). Considering this evidence in humans, an intriguing future perspective will be to explore the existence of clinical cases linked to increased DDO gene expression (i.e. DDO copy number variations) and evaluate their possible association with neurodevelopmental disorders, using advanced technologies tools (Iossa et al. 2015; Zebisch et al. 2015). On the other hand, the absence of dramatic alterations in adult mutant mice developed with trace levels of cerebral d-Asp could be the result of unknown adaptive events triggered over lifetime to counteract the lack of this d-amino acid. Future studies are mandatory to investigate the neurobiological effect of d-Asp depletion at earlier developmental stages, when eventual adaptive events could be not yet established.
Our behavioral results revealed that Ddo knockin mice exhibit a paradoxical cognitive enhancement. Indeed, we reported improved object recognition and spatial memory abilities in Ddo knockin mice. To our knowledge, this phenotype is hardly attributable to d-Asp depletion since we and others have previously found similar cognitive improvement in young-adult Ddo knockout animals, characterized by congenital increased levels of d-Asp, and in rodents exogenously administered with this d-amino acid (Errico et al. 2008b, 2011a, b; Topo et al. 2010). On the other hand, we cannot exclude that the cognitive changes found in Ddo knockin mice may result from the appearance of a still unknown secondary function for DDO, which is manifested in mutant mice only under a condition of precocious DDO overexpression. Consistently, it is important to remark that DDO activity is up to 100 times higher in the neonatal brain of R26Ddo/Ddo mice, compared to age-matched control littermates. Nevertheless, at this stage of knowledge, the hypothesis of a gain of function for DDO is only speculative and, therefore, new studies are warranted to clarify the origin of the unexpected cognitive phenotype found in Ddo knockin mice. Interestingly, while the Ddo transcription levels were overall stably overexpressed with age, DDO activity significantly decreased in a time-dependent manner, in both R26Ddo/+ and R26 Ddo/Ddo whole brains. Unfortunately, the lack of available selective anti-DDO antibodies does not allow us to quantify the DDO protein amount and, in turn, to discriminate the specific mechanisms/factors involved in the time-dependent inhibition of DDO activity found in Ddo knockin brains.
Previous findings revealed a close relationship between glutamatergic transmission, PV-positive interneurons and cognitive performance (Ferguson and Gao 2018a, b; Rossi et al. 2012). Indeed, it has been established that a reduced number of these connection neurons affects cortical excitatory activity of glutamatergic pyramidal neurons and produces cognitive impairments in mice, leading to phenotypes resembling those observed in psychiatric disorders, including schizophrenia (Ferguson and Gao 2018a). However, as PV interneurons are important regulators of excitatory/inhibitory balance in the PFC, disruption of such balance in either direction can alter cognitive abilities and social interaction (Ferguson and Gao 2018b; Rossi et al. 2012). Based on these considerations, future electrophysiological studies will help to clarify whether the cognitive alterations observed in Ddo knockin mice are functionally linked to changes in the activity of cortical PV-positive interneurons.
In conclusion, we report for the first time that early d-Asp removal in mice by precocious DDO overexpression induced selective neuronal and cognitive phenotypes that are overall counterintuitive to those expected (Errico et al. 2018; Hashimoto et al. 1993; Sakai et al. 1998; Wolosker et al. 2000). On the other hand, the present observations confirm the influence of physiological time-dependent DDO expression and d-Asp occurrence in regulating the correct brain formation and functioning.
References
Cristino L, Luongo L, Squillace M, Paolone G, Mango D, Piccinin S, Zianni E, Imperatore R, Iannotta M, Longo F, Errico F, Vescovi AL, Morari M, Maione S, Gardoni F, Nistico R, Usiello A (2015) d-Aspartate oxidase influences glutamatergic system homeostasis in mammalian brain. Neurobiol Aging 36(5):1890–1902. https://doi.org/10.1016/j.neurobiolaging.2015.02.003
Cuomo M, Keller S, Punzo D, Nuzzo T, Affinito O, Coretti L, Carella M, de Rosa V, Florio E, Boscia F, Avvedimento VE, Cocozza S, Errico F, Usiello A, Chiariotti L (2019) Selective demethylation of two CpG sites causes postnatal activation of the Dao gene and consequent removal of d-serine within the mouse cerebellum. Clin Epigenet 11(1):149. https://doi.org/10.1186/s13148-019-0732-z
D'Aniello A, Vetere A, Petrucelli L (1993) Further study on the specificity of d-amino acid oxidase and d-aspartate oxidase and time course for complete oxidation of d-amino acids. Comp Biochem Physiol B 105(3–4):731–734
de Bartolomeis A, Tomasetti C (2012) Calcium-dependent networks in dopamine-glutamate interaction: the role of postsynaptic scaffolding proteins. Mol Neurobiol 46(2):275–296. https://doi.org/10.1007/s12035-012-8293-6
de Rosa V, Secondo A, Pannaccione A, Ciccone R, Formisano L, Guida N, Crispino R, Fico A, Polishchuk R, D'Aniello A, Annunziato L, Boscia F (2019) D-Aspartate treatment attenuates myelin damage and stimulates myelin repair. EMBO Mol Med. https://doi.org/10.15252/emmm.201809278
Di Giorgi-Gerevini V, Melchiorri D, Battaglia G, Ricci-Vitiani L, Ciceroni C, Busceti CL, Biagioni F, Iacovelli L, Canudas AM, Parati E, De Maria R, Nicoletti F (2005) Endogenous activation of metabotropic glutamate receptors supports the proliferation and survival of neural progenitor cells. Cell Death Differ 12(8):1124–1133. https://doi.org/10.1038/sj.cdd.4401639
Dunlop DS, Neidle A, McHale D, Dunlop DM, Lajtha A (1986) The presence of free d-aspartic acid in rodents and man. Biochem Biophys Res Commun 141(1):27–32
Errico F, Pirro MT, Affuso A, Spinelli P, De Felice M, D'Aniello A, Di Lauro R (2006) A physiological mechanism to regulate D -aspartic acid and NMDA levels in mammals revealed by d-aspartate oxidase deficient mice. Gene 374:50–57
Errico F, Nistico R, Palma G, Federici M, Affuso A, Brilli E, Topo E, Centonze D, Bernardi G, Bozzi Y, D'Aniello A, Di Lauro R, Mercuri NB, Usiello A (2008a) Increased levels of d-aspartate in the hippocampus enhance LTP but do not facilitate cognitive flexibility. Mol Cell Neurosci 37(2):236–246. https://doi.org/10.1016/j.mcn.2007.09.012
Errico F, Rossi S, Napolitano F, Catuogno V, Topo E, Fisone G, D'Aniello A, Centonze D, Usiello A (2008b) d-aspartate prevents corticostriatal long-term depression and attenuates schizophrenia-like symptoms induced by amphetamine and MK-801. J Neurosci 28(41):10404–10414. https://doi.org/10.1523/JNEUROSCI.1618-08.2008
Errico F, Nistico R, Napolitano F, Mazzola C, Astone D, Pisapia T, Giustizieri M, D'Aniello A, Mercuri NB, Usiello A (2011a) Increased d-aspartate brain content rescues hippocampal age-related synaptic plasticity deterioration of mice. Neurobiol Aging 32(12):2229–2243. https://doi.org/10.1016/j.neurobiolaging.2010.01.002
Errico F, Nistico R, Napolitano F, Oliva AB, Romano R, Barbieri F, Florio T, Russo C, Mercuri NB, Usiello A (2011b) Persistent increase of d-aspartate in d-aspartate oxidase mutant mice induces a precocious hippocampal age-dependent synaptic plasticity and spatial memory decay. Neurobiol Aging 32(11):2061–2074. https://doi.org/10.1016/j.neurobiolaging.2009.12.007
Errico F, Nistico R, Di Giorgio A, Squillace M, Vitucci D, Galbusera A, Piccinin S, Mango D, Fazio L, Middei S, Trizio S, Mercuri NB, Teule MA, Centonze D, Gozzi A, Blasi G, Bertolino A, Usiello A (2014) Free d-aspartate regulates neuronal dendritic morphology, synaptic plasticity, gray matter volume and brain activity in mammals. Transl Psychiatry 4:e417. https://doi.org/10.1038/tp.2014.59
Errico F, Nuzzo T, Carella M, Bertolino A, Usiello A (2018) The emerging role of altered d-aspartate metabolism in schizophrenia: new insights from preclinical models and human studies. Front Psychiatry 9:559. https://doi.org/10.3389/fpsyt.2018.00559
Fagg GE, Matus A (1984) Selective association of N-methyl aspartate and quisqualate types of l-glutamate receptor with brain postsynaptic densities. Proc Natl Acad Sci USA 81(21):6876–6880
Ferguson BR, Gao WJ (2018a) PV interneurons: critical regulators of E/I balance for prefrontal cortex-dependent behavior and psychiatric disorders. Front Neural Circ 12:37. https://doi.org/10.3389/fncir.2018.00037
Ferguson BR, Gao WJ (2018b) Thalamic control of cognition and social behavior via regulation of gamma-aminobutyric acidergic signaling and excitation/inhibition balance in the medial prefrontal cortex. Biol Psychiatry 83(8):657–669. https://doi.org/10.1016/j.biopsych.2017.11.033
Gong XQ, Frandsen A, Lu WY, Wan Y, Zabek RL, Pickering DS, Bai D (2005) D-aspartate and NMDA, but not l-aspartate, block AMPA receptors in rat hippocampal neurons. Br J Pharmacol 145(4):449–459. https://doi.org/10.1038/sj.bjp.0706199
Hashimoto A, Kumashiro S, Nishikawa T, Oka T, Takahashi K, Mito T, Takashima S, Doi N, Mizutani Y, Yamazaki T et al (1993) Embryonic development and postnatal changes in free d-aspartate and d-serine in the human prefrontal cortex. J Neurochem 61(1):348–351
Hashimoto A, Oka T, Nishikawa T (1995) Anatomical distribution and postnatal changes in endogenous free d-aspartate and d-serine in rat brain and periphery. Eur J Neurosci 7(8):1657–1663
Horio M, Ishima T, Fujita Y, Inoue R, Mori H, Hashimoto K (2013) Decreased levels of free d-aspartic acid in the forebrain of serine racemase (Srr) knock-out mice. Neurochem Int 62(6):843–847. https://doi.org/10.1016/j.neuint.2013.02.015
Ikonomidou C (2009) Triggers of apoptosis in the immature brain. Brain Dev 31(7):488–492. https://doi.org/10.1016/j.braindev.2009.02.006
Iossa S, Costa V, Corvino V, Auletta G, Barruffo L, Cappellani S, Ceglia C, Cennamo G, D'Adamo AP, D'Amico A, Di Paolo N, Forte R, Gasparini P, Laria C, Lombardo B, Malesci R, Vitale A, Marciano E, Franze A (2015) Phenotypic and genetic characterization of a family carrying two Xq21.1–21.3 interstitial deletions associated with syndromic hearing loss. Mol Cytogenet 8:18. https://doi.org/10.1186/s13039-015-0120-0
Ito T, Hayashida M, Kobayashi S, Muto N, Hayashi A, Yoshimura T, Mori H (2016) Serine racemase is involved in d-aspartate biosynthesis. J Biochem. https://doi.org/10.1093/jb/mvw043
Jagannath V, Marinova Z, Monoranu CM, Walitza S, Grunblatt E (2017) Expression of d-amino acid oxidase (DAO/DAAO) and d-amino acid oxidase activator (DAOA/G72) during development and aging in the human post-mortem brain. Front Neuroanat 11:31. https://doi.org/10.3389/fnana.2017.00031
Jansson LC, Akerman KE (2014) The role of glutamate and its receptors in the proliferation, migration, differentiation and survival of neural progenitor cells. J Neural Transm (Vienna) 121(8):819–836. https://doi.org/10.1007/s00702-014-1174-6
Katane M, Homma H (2010) d-aspartate oxidase: the sole catabolic enzyme acting on free d-aspartate in mammals. Chem Biodivers 7(6):1435–1449. https://doi.org/10.1002/cbdv.200900250
Katane M, Kanazawa R, Kobayashi R, Oishi M, Nakayama K, Saitoh Y, Miyamoto T, Sekine M, Homma H (2017) Structure-function relationships in human d-aspartate oxidase: characterisation of variants corresponding to known single nucleotide polymorphisms. Biochim Biophys Acta Proteins Proteom 1865(9):1129–1140. https://doi.org/10.1016/j.bbapap.2017.06.010
Katane M, Kuwabara H, Nakayama K, Saitoh Y, Miyamoto T, Sekine M, Homma H (2018) Rat d-aspartate oxidase is more similar to the human enzyme than the mouse enzyme. Biochim Biophys Acta Proteins Proteom 1866(7):806–812. https://doi.org/10.1016/j.bbapap.2017.12.009
Kitamura A, Hojo Y, Ikeda M, Karakawa S, Kuwahara T, Kim J, Soma M, Kawato S, Tsurugizawa T (2018) Ingested d-aspartate facilitates the functional connectivity and modifies dendritic spine morphology in rat hippocampus. Cereb Cortex. https://doi.org/10.1093/cercor/bhy120
Lim L, Mi D, Llorca A, Marin O (2018) Development and functional diversification of cortical interneurons. Neuron 100(2):294–313. https://doi.org/10.1016/j.neuron.2018.10.009
Molinaro G, Pietracupa S, Di Menna L, Pescatori L, Usiello A, Battaglia G, Nicoletti F, Bruno V (2010) D-aspartate activates mGlu receptors coupled to polyphosphoinositide hydrolysis in neonate rat brain slices. Neurosci Lett 478(3):128–130. https://doi.org/10.1016/j.neulet.2010.04.077
Molla G, Chaves-Sanjuan A, Savinelli A, Nardini M, Pollegioni L (2020) Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain. FASEB J 34(1):1182–1197. https://doi.org/10.1096/fj.201901703R
Neidle A, Dunlop DS (1990) Developmental changes in free d-aspartic acid in the chicken embryo and in the neonatal rat. Life Sci 46(21):1517–1522
Nicoletti CG, Monteleone F, Marfia GA, Usiello A, Buttari F, Centonze D, Mori F (2019) Oral d-Aspartate enhances synaptic plasticity reserve in progressive multiple sclerosis. Mult Scler. https://doi.org/10.1177/1352458519828294
Nuzzo T, Sacchi S, Errico F, Keller S, Palumbo O, Florio E, Punzo D, Napolitano F, Copetti M, Carella M, Chiariotti L, Bertolino A, Pollegioni L, Usiello A (2017) Decreased free d-aspartate levels are linked to enhanced d-aspartate oxidase activity in the dorsolateral prefrontal cortex of schizophrenia patients. NPJ Schizophr 3:16. https://doi.org/10.1038/s41537-017-0015-7
Nuzzo T, Feligioni M, Cristino L, Pagano I, Marcelli S, Iannuzzi F, Imperatore R, D'Angelo L, Petrella C, Carella M, Pollegioni L, Sacchi S, Punzo D, De Girolamo P, Errico F, Canu N, Usiello A (2019a) Free d-aspartate triggers NMDA receptor-dependent cell death in primary cortical neurons and perturbs JNK activation, Tau phosphorylation, and protein SUMOylation in the cerebral cortex of mice lacking d-aspartate oxidase activity. Exp Neurol 317:51–65. https://doi.org/10.1016/j.expneurol.2019.02.014
Nuzzo T, Punzo D, Devoto P, Rosini E, Paciotti S, Sacchi S, Li Q, Thiolat ML, Vega C, Carella M, Carta M, Gardoni F, Calabresi P, Pollegioni L, Bezard E, Parnetti L, Errico F, Usiello A (2019b) The levels of the NMDA receptor co-agonist d-serine are reduced in the substantia nigra of MPTP-lesioned macaques and in the cerebrospinal fluid of Parkinson's disease patients. Sci Rep 9(1):8898. https://doi.org/10.1038/s41598-019-45419-1
Olverman HJ, Jones AW, Mewett KN, Watkins JC (1988) Structure/activity relations of N-methyl-d-aspartate receptor ligands as studied by their inhibition of [3H]d-2-amino-5-phosphonopentanoic acid binding in rat brain membranes. Neuroscience 26(1):17–31. https://doi.org/10.1016/0306-4522(88)90124-8
Paxinos G, Franklin KBJ (2001) The mouse brain in stereotaxic coordinates, 2nd edn. Academic, New York
Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G (2007) Physiological functions of d-amino acid oxidases: from yeast to humans. Cell Mol Life Sci 64(11):1373–1394. https://doi.org/10.1007/s00018-007-6558-4
Punzo D, Errico F, Cristino L, Sacchi S, Keller S, Belardo C, Luongo L, Nuzzo T, Imperatore R, Florio E, De Novellis V, Affinito O, Migliarini S, Maddaloni G, Sisalli MJ, Pasqualetti M, Pollegioni L, Maione S, Chiariotti L, Usiello A (2016) Age-related changes in d-aspartate oxidase promoter methylation control extracellular d-aspartate levels and prevent precocious cell death during brain aging. J Neurosci 36(10):3064–3078. https://doi.org/10.1523/JNEUROSCI.3881-15.2016
Querques F, Cantilena B, Cozzolino C, Esposito MT, Passaro F, Parisi S, Lombardo B, Russo T, Pastore L (2015) Angiotensin receptor I stimulates osteoprogenitor proliferation through TGFbeta-mediated signaling. J Cell Physiol 230(7):1466–1474. https://doi.org/10.1002/jcp.24887
Rossi MA, Hayrapetyan VY, Maimon B, Mak K, Je HS, Yin HH (2012) Prefrontal cortical mechanisms underlying delayed alternation in mice. J Neurophysiol 108(4):1211–1222. https://doi.org/10.1152/jn.01060.2011
Sacchi S, Novellis V, Paolone G, Nuzzo T, Iannotta M, Belardo C, Squillace M, Bolognesi P, Rosini E, Motta Z, Frassineti M, Bertolino A, Pollegioni L, Morari M, Maione S, Errico F, Usiello A (2017) Olanzapine, but not clozapine, increases glutamate release in the prefrontal cortex of freely moving mice by inhibiting d-aspartate oxidase activity. Sci Rep 7:46288. https://doi.org/10.1038/srep46288
Sakai K, Homma H, Lee JA, Fukushima T, Santa T, Tashiro K, Iwatsubo T, Imai K (1998) Emergence of d-aspartic acid in the differentiating neurons of the rat central nervous system. Brain Res 808(1):65–71
Silberschmidt D, Rodriguez-Mallon A, Mithboakar P, Cali G, Amendola E, Sanges R, Zannini M, Scarfo M, De Luca P, Nitsch L, Di Lauro R, De Felice M (2011) In vivo role of different domains and of phosphorylation in the transcription factor Nkx2-1. BMC Dev Biol 11:9. https://doi.org/10.1186/1471-213X-11-9
Tanaka-Hayashi A, Hayashi S, Inoue R, Ito T, Konno K, Yoshida T, Watanabe M, Yoshimura T, Mori H (2014) Is d-aspartate produced by glutamic-oxaloacetic transaminase-1 like 1 (Got1l1): a putative aspartate racemase? Amino Acids. https://doi.org/10.1007/s00726-014-1847-3
Topo E, Soricelli A, Di Maio A, D'Aniello E, Di Fiore MM, D'Aniello A (2010) Evidence for the involvement of d-aspartic acid in learning and memory of rat. Amino Acids 38(5):1561–1569. https://doi.org/10.1007/s00726-009-0369-x
Tremblay R, Lee S, Rudy B (2016) GABAergic interneurons in the neocortex: from cellular properties to circuits. Neuron 91(2):260–292. https://doi.org/10.1016/j.neuron.2016.06.033
Van Veldhoven PP, Brees C, Mannaerts GP (1991) D-aspartate oxidase, a peroxisomal enzyme in liver of rat and man. Biochim Biophys Acta 1073(1):203–208
Wang LZ, Zhu XZ (2003) Spatiotemporal relationships among d-serine, serine racemase, and d-amino acid oxidase during mouse postnatal development. Acta Pharmacol Sin 24(10):965–974
Wolosker H, Sheth KN, Takahashi M, Mothet JP, Brady RO Jr, Ferris CD, Snyder SH (1999) Purification of serine racemase: biosynthesis of the neuromodulator d-serine. Proc Natl Acad Sci USA 96(2):721–725
Wolosker H, D'Aniello A, Snyder SH (2000) D-aspartate disposition in neuronal and endocrine tissues: ontogeny, biosynthesis and release. Neuroscience 100(1):183–189
Yamada R, Nagasaki H, Wakabayashi Y, Iwashima A (1988) Presence of d-aspartate oxidase in rat liver and mouse tissues. Biochim Biophys Acta 965(2–3):202–205. https://doi.org/10.1016/0304-4165(88)90057-8
Zebisch A, Schulz E, Grosso M, Lombardo B, Acierno G, Sill H, Iolascon A (2015) Identification of a novel variant of epsilon-gamma-delta-beta thalassemia highlights limitations of next generation sequencing. Am J Hematol 90(3):E52–54. https://doi.org/10.1002/ajh.23913
Acknowledgements
We thank Massimo Pasqualetti and Mehdi Eshraghi for their helpful discussion and comments.
Funding
This work was supported by a grant from MIUR (Ministero dell’Istruzione, dell’Università e della Ricerca, Progetto PRIN 2017—Project nr 2017M42834) to A.U.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All research involving animals was carried out in accordance with the Italian directive of the Ministry of Health governing animal welfare and protection (D.LGS26/2014), and approved by “Direzione Generale della Sanità e dei Farmaci Veterinari (Ufficio 6)” (permission nr 796/2018).
Informed consent
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Handling editor: V. Bolshakov.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
De Rosa, A., Mastrostefano, F., Di Maio, A. et al. Prenatal expression of d-aspartate oxidase causes early cerebral d-aspartate depletion and influences brain morphology and cognitive functions at adulthood. Amino Acids 52, 597–617 (2020). https://doi.org/10.1007/s00726-020-02839-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-020-02839-y