Abstract
Polyamines (PAs), such as putrescine, spermidine and spermine, are alkyl-amines that are essential for cell growth, proliferation, differentiation and cancer progression in eukaryotic cells. A designed PA analogue; DENSpm, induces cell cycle arrest, inhibits proliferation and induces apoptosis in melanoma, breast, prostate, lung and colon cancer cells. Although the mechanism by which DENSpm induces apoptosis has been examined, the effect of DENSpm on autophagy has not been investigated yet. Therefore, in this study, our objective was to determine the role of p53 in the DENSpm-induced autophagy/apoptotic regulation in a time-dependent manner in colon cancer cells. Exposure of HCT 116 colon cancer cells to DENSpm decreased cell viability in a dose- and time-dependent manner. However, the p53 mutant, SW480, and deficient HCT 116 p53−/− cells were more resistant to DENSpm treatment compared to HCT 116 p53+/+ cells. The resistant profile caused by p53 defect also caused a cell type-specific response to PA pool depletion and SSAT overexpression. In addition to PA depletion, DENSpm induced apoptosis by activating the mitochondria-mediated pathway in a caspase-dependent manner regardless of p53 expression in colon cancer cells. Concomitantly, we determined that DENSpm also affected autophagy in HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon cancer cells for different periods of exposure to DENSpm. Therefore, this study revealed that effect of DENSpm on cell death differs due to p53 protein expression profile. In addition, DENSpm-induced autophagy may be critical in drug resistance in colon cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyamines (PAs) are amine-derived organic cations, present in all organisms (Cohen 1978). Putrescine (Put), spermidine (Spd) and spermine (Spm), are natural PAs with essential roles in cell proliferation, growth, differentiation and cancer progression (Igarashi and Kashiwagi 2000). Because high intracellular PA levels are associated with cancer development and progression, chemotherapeutics that activate PA catabolism have gained importance as a new anticancer therapy model (Pegg 1988). The PA analogues, N1-ethyl-N11-((cycloheptyl) methyl)-4,8-diazaundecane (CHENSpm) and N1, N11, diethylnorspermine (DENSpm) which were synthesized to decrease the intracellular PA pool (Saab et al. 1993), were investigated for their growth inhibitory potential in different cancer cells (Ha et al. 1997).
DENSpm is one of the most studied PA analogues, and induces cell cycle arrest at the G1 phase in MALME-3 M human melanoma cells by targeting the p53-p21-Rb signaling axis (Kramer et al. 1999). A recent report showed that DENSpm treatment at cell death inducing concentrations caused increased activity of the spermidine-spermine acetyl transferase (SSAT) and apoptotic responses, such as caspase-3 cleavage and cytochrome c release from mitochondria in SK-MEL-28 melanoma cells (Chen et al. 2001). Similarly, apoptotic induction caused by DENSpm treatment was shown in breast (Hegardt et al. 2002), non-small lung (Hahm et al. 2002) and prostate cancer cells (Zagaja et al. 1998). DENSpm was used in Phase I and II clinical trials, but the drug efficiency was found insignificant in breast and prostate cancer patients (Schipper et al. 2000; Wolff et al. 2003). The potential effect of DENSpm on the DNA damaging agents; oxaliplatin or cisplatin was promising for ovarian cancer cells (Hector et al. 2004). Similarly, it was suggested that DENSpm might synergize with apoptotic efficiency of 5-FU (5-fluorouracil) in colon cancer cells (Allen et al. 2007). Although the molecular players of DENSpm-induced cell death are not fully identified, the utilization of DENSpm in combination therapy models may be beneficial compared to single agent treatment models.
Autophagy is an evolutionarily conserved mechanism important for balancing sources of energy at critical conditions such as during development, nutrient deprivation or inflammatory stress. Autophagy is characterized as the self-degradation of cellular parts by eliminating misfolded or aggregated proteins and damaged organelles, such as mitochondria, endoplasmic reticulum, peroxisomes and intracellular pathogens (Mizushima 2009). Autophagy process is generally initiated by the surrounding of intracellular content with a crescent-shaped membrane (phagophore) that is maintained to form a closed double-membrane, called the autophagosomes. The finalization of autophagy with prolonged stimuli leads to the fusion of the autophagosome and lysosome to form an autolysosome (Yang and Klionsky 2009). One of the essential autophagy molecules is Beclin-1, with structural similarity to the Bcl-2 homology (BH) 3 domain found in pro-apoptotic Bcl-2 family members. Therefore, Beclin-1 is proposed as a linker protein between apoptosis and autophagy because of its interaction with anti-apoptotic Bcl-2 family members, and it is suggested that overexpression of Bcl-2 may prevent autophagy when cells are exposed to death inducers (Pattingre and Levine 2006).
The complex relationship between autophagy and apoptosis in cancer cells was demonstrated by studies on tumor suppressor protein, p53 (Crighton et al. 2006). p53 transcribes the pro-apoptotic Bcl-2 family proteins, Bax, Puma and Noxa, which leads to mitochondria-mediated apoptosis via oligomerization of Bax following apoptotic stimuli (Morselli et al. 2008a). In addition to apoptotic cell death, the functional role of p53 and molecular mechanism of p53 during autophagy process was determined. Whereas nuclear localization of p53 acts as an autophagy-promoting transcription factor, cytoplasmic state of p53 may inhibit autophagy. Thus, p53 is present in mutant forms in the majority of cancer cells, which modulate the therapeutic efficiency of drugs, inducing both apoptosis and autophagy.
In this study, our objective was to investigate the potential effect of DENSpm on autophagy in colon cancer cells that express functionally different p53 protein. We found that the PA analogue DENSpm caused a significant increase in the apoptotic cell population by activating the mitochondria-mediated pathway in HCT 116 p53+/+ colon cancer cells. However, the p53 mutant SW480 and p53-deficient HCT 116 p53−/− colon cancer cells showed resistance to DENSpm-activated apoptosis due to the induction of autophagy. In summary, we propose a potential axis of autophagy and apoptosis induced by DENSpm in a p53-dependent manner in colon cancer cells. Understanding of this regulation may help to develop autophagy-based therapeutic interventions for colon cancer.
Materials and methods
Drugs, chemical and antibodies
DENSpm was purchased from TOCRIS (Tocris Bioscience, UK), dissolved in water to prepare a 10-mM stock solution and aliquots were stored at −20 °C. Acridine orange (AO) was purchased from Sigma (St. Louis, MO, USA), dissolved in DMSO (dimethylsulfoxide) to prepare a 10-mM stock solution, and aliquots were stored at −20 °C. β-Actin, PARP, pro-caspase-9, pro-caspase-7, pro-caspase-3, Bcl-xL, Mcl-1, Puma, Atg3, Atg5, Atg7, Atg12 and LC3 (1:1,000 dilution each) rabbit antibodies were purchased from cell signaling technology (CST, Danvers, MA, USA). SSAT, PAO (1:1,000 dilution each) rabbit antibodies were purchased from Santa Cruz (Santa Cruz, CA, USA). ODC and AZI were a kind gift from Chaim Kahana (Weizmann Institute, Rehovot, Israel). Beclin-1, Bcl-2, Bax and p62 (1:1,000 dilution each) mouse antibodies were purchased from Becton–Dickinson Biosciences (BD Biosciences, Bedford, MA). HRP-conjugated secondary anti-rabbit and anti-mouse antibodies (1:5,000) were from CST (Danvers, MA, USA).
Cell lines and culture conditions
HCT 116 (CCL-247) and SW480 (CCL-228) cells were purchased from the American Type Culture Collection (ATCC, Manassas, USA). HCT 116 p53−/− human colon cancer cells were kindly provided by Batu Erman (Sabancı University, Tuzla, Turkey). HCT 116, HCT 116 p53−/− and SW480 colon cancer cells were maintained in McCoy’s medium (PAN Biotech, Aidenbach, Germany) and MEM (PAN Biotech, Aidenbach, Germany), respectively, with 2 mM l-glutamine, 10 % fetal calf serum (PAN Biotech), 1 % non-essential amino acids (Biological Industries, Kibbutz Beit-Haemek, Israel), and penicillin (100 U/ml)- streptomycin (100 μg/ml) (Biological Industries, Kibbutz Beit-Haemek, Israel) and grown in the presence of 5 % CO2 in humidified air at 37 °C.
MTT cell viability assay
The dose-dependent effect of DENSpm on cell viability was determined by colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide [(MTT), Roche, Indianapolis, IN, USA)] assay in HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon cancer cells. Each cell line was seeded at 1 × 104 cells/well in 96 well plates and treated with various concentrations of DENSpm (0–50 μM) for 24, 48 and 72 h. Following exposure of the cells to DENSpm, 10-μl MTT dye [5 mg/ml in 1× PBS (Phosphate-buffered saline), Sigma (St. Louis, MO, USA)] was added to the wells and then incubated at 37 °C for 4 h for conversion of MTT to formazan crystals by the activation of mitochondrial enzymes of viable cells. Following aspiration of the medium, 200-μl DMSO (Sigma; St. Louis, MO, USA) was added, and the absorbance of the suspension was determined at 570 nm with a microplate reader (Bio-Rad, Hercules, CA, USA).
Cell cycle analysis by PI staining
HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon carcinoma cells at a density of 2 × 105 cells/well were seeded in 6 well plates, and then treated with DENSpm (10 μM) for 24, 48 and 72 h. Both floating and adherent cells were collected and fixed with 70 % ethanol. Following incubation on ice for 30 min, the samples were centrifuged at 1,200 rpm for 5 min. The pellets were resuspended in 1× PBS with RNase (100 μg/ml) and propidium iodide (PI) solution (40 μg/ml). The samples were incubated for 30 min at 37 °C in the dark and then analyzed on Accuri C6 Flow Cytometer (BD Biosciences, Bedford, MA), equipped with a 15 mW, 488 nm, air cooled argon ion laser. Fluorescence emission was collected through a 570 nm band-pass filter. Cell cycle distribution was performed using the FlowJo software (BD Biosciences, Bedford, MA).
PA analysis by HPLC
The intracellular PA content was determined by HPLC analysis following a benzoylation procedure. HCT 116 p53+/+, SW480 and HCT 116 p53−/− (1.2 × 106) cells were seeded in 100-mm petri dishes and allowed to attach overnight. The cells were then treated with the desired concentration of DENSpm for 24, 48 and 72 h. Following a washing step with 1× PBS, the cell lysates were obtained by scraping. The scraped cell lysates were transferred to a new microfuge tube and 50 % trichloroacetic acid was added to each sample (1:10, v/v). All samples were stored at −20 °C until the benzoylation process. Following benzoylation, the samples were immediately analyzed on HPLC system (Agilent, Santa Clara, CA, USA) using a UV detector set at 226 mV. The results were compared to the internal standard 1,7-diaminoheptane and the standard curve for Put, Spd, and Spm standards (10-mM stock concentration each).
Determination of loss of mitochondrial membrane potential (ΔΨm)
We measured the Δψm loss in HCT 116 p53+/+, SW480 and HCT 116 p53−/− cells using 5, 5′,6, 6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocynanine iodide (JC-1; (MitoScreen Flow Cytometer Mitochondrial Membrane Potential Detection Kit, BD Biosciences, Bedford, MA). A total of 2 × 105 cells/well were seeded in 12 well plates, allowed to attached overnight, and then treated with desired concentrations of DENSpm (10 μM) for 24, 48 and 72 h. Following a washing step with 1× PBS, the cells were resuspendend in JC-1 working solution for 10 min at 37 °C in CO2 incubator. After washing cells gently with 1× Assay Buffer, cells were centrifuged at 400×g for 5 min and resuspended in 0.5 1× Assay Buffer. DENSpm-induced Δψm disruption in time-dependent manner was analyzed by FACS Flow cytometer (BD Biosciences, Bedford, MA). Untreated cells with healthy mitochondria were detected by measuring the increase in the red fluorescence (FL-2 channel) due to JC-1 aggregates. The cells that have depolarized Δψm due to DENSpm treatment emitting JC-1 monomers were detected by increase in the green fluorescence (FL-1 channel).
Detection of intracellular reactive oxygen species (ROS) generation
HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon cancer cells were plated at a density of 2.5 × 105 cells per well in 6 well plates. Following exposure of cells to DENSpm (10 μM) for 24, 48, and 72 h, the cells were harvested by trypsinization. The pellets were resuspendend in 1× PBS with 10 μM of CM-H2DCFDA, and the cells incubated in a cell incubator [(37 °C), high relative humidity (95 %), and controlled CO2 level (5 %)] in the dark for 45 min. A total of 5,000 events were analyzed in FACS flow cytometer (Accuri C6 Flow Cytometer, BD Biosciences, Bedford, MA). The fluorescence level of the CM-H2DCFDA staining was analyzed on the FL-1 channel (525 nm).
Protein extraction and immunoblotting
HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon carcinoma cells were treated with the appropriate concentrations of DENSpm for 24, 48, and 72 h. First, all samples were washed with ice-cold 1× PBS and lysed on ice in a solution containing 20-mM Tris-HCl (pH 7.5), 150-mM NaCl, 0.5 % Nonidet P-40 (v/v), 1-mM EDTA, 0.5-mM PMSF, 1-mM DTT and protease inhibitor cocktail (Roche, Indianapolis, IN, USA). After cell lysis, the cell debris was removed by the centrifugation for 15 min at 13,200 rpm, and the protein concentrations were determined by Bradford protein assay (Bio-Rad, Hercules, CA, USA). The total protein lysates (30 μg) were separated by a 12 % SDS-PAGE (Sodium dodecyl sulphate polyacrylamide gel electrophoresis) and transferred onto PVDF (Polyvinyldifluoride) membranes (Roche, Indianapolis, IN, USA). The membranes were then blocked with 5 % milk blocking solution in Tris buffered saline (TBS)-Tween-20 (Sigma, St. Louis, MO, USA) and incubated with the appropriate primary and horseradish peroxidase (HRP)-conjugated secondary antibodies (CST, Danvers, MA, USA) in antibody buffer containing 5 % (v/v) milk blocking solution. Following gentle washing step with 1× TBS-Tween20, the proteins were analyzed using an enhanced ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA, USA).
Autophagic vacuole formation assays
Acridine orange (AO) staining
HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon cancer cells were seeded at a density of 1 × 105 cells/well in 6 well plates. Following exposure of cells to DENSpm (10 μM) for 24, 48, and 72 h, the samples were treated with AO solution (5 μg/ml) for 15 min at 37 °C. After dye incubation, the media were carefully discarded and the cells were collected in 1× PBS. To eliminate the DENSpm-induced apoptotic cells, resuspended cells were stained by PI (1 μg/ml) and immediately 10,000 cells per sample was analyzed with a FACS flow cytometer (Attun NxT Acoustic Focusing Cytometer, Applied Biosciences, MA USA) using the FL-1 (green fluorescence) and FL-3 channel (red fluorescence). The decrease in green fluorescence levels was accepted as autophagy vacuoles through gating cells according to PI-stained untreated cells.
Determination of DENSpm-induced GFP-LC3 dot formation by flow cytometer
HCT 116, SW480 and HCT 116 p53−/− colon carcinoma cells were seeded in 6-well plates and transfected with a green fluorescent protein (GFP)-tagged LC3 expressing vector (0.5 μg/ml) using the FuGENE6 (Promega, Sunnyvale, CA, USA). After 24 h transfection, the cells were exposed to DENSpm (10 µM) for 24, 48, and 72 h. Following DENSpm treatment, 10,000 GFP-LC3 transfected cells per sample were analyzed with flow cytometer using the FL-1 channel (green fluorescence) (Accuri C6 Flow Cytometer (BD Biosciences, Bedford, MA).
Statistical analysis
All the experiments were statistically analyzed by a two-way ANOVA using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). Statistically significant results by ANOVA were further analyzed by Bonferroni post hoc analysis (where indicated). A p value <0.05 was considered statistically significant. Error bars in the graphs were generated using the ± standard deviation (SD) values. The immunoblotting results were repeated at least twice and the Image J program was used to obtain the band intensities.
Results
DENSpm treatment prevented proliferation and cell survival in time-dependent manner
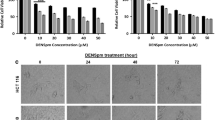
To understand the cytotoxic effect of DENSpm in p53 wt (HCT 116 p53+/+), p53 mutant (SW480) and knockout (HCT 116 p53−/−) colon cancer cells, MTT cell viability assay was performed following treatment with various concentrations of DENSpm (0–50 µM) at different time points. DENSpm induced loss of cell viability in dose- and time-dependent manner in HCT 116 p53+/+, SW480 and HCT 116 p53−/− colon cancer cells (Fig. 1a–c). Exposure of HCT 116 p53+/+ cells treated with increasing concentrations of DENSpm had the largest decrease in cell viability compared to p53 deficient HCT 116 colon cancer cells. Based on the MTT cell viability assay results, 10-μM DENSpm treatment decreased cell viability by 15, 35 and 50 % compared to the untreated control HCT 116 p53+/+cells at 24, 48, and 72 h, respectively (Fig. 1a; ***p < 0.0001). Although the loss of cell viability caused by DENSpm for 24 h was less efficient in SW480 and HCT 116 p53−/− colon cancer cells (11 and 9 % vs control untreated cells for each cell lines, respectively, *p < 0.05), long-term exposure of these cells to DENSpm for 48 and 72 h further decreased cell viability by 24 and 32 % in SW480, 17 and 24 % in HCT 116 p53−/− colon cancer cells, respectively (Fig. 1b, c). According to time- and dose-dependent MTT cell viability assay results, DENSpm-induced cell viability loss may be due to enhanced cell death or reduction of cell viability in colon cancer cells. 10-μM DENSpm was chosen and used in further experiments to evaluate the efficiency of DENSpm in colon cancer cells. To confirm MTT results, the effect of DENSpm on cell viability was also determined by trypan blue dye exclusion assay. Although the suppressive effect of DENSpm on cell viability was determined in HCT 116 p53+/+ and SW480 colon cancer cells in a time-dependent manner, DENSpm did not cause a cytotoxic effect in HCT 116 p53−/− colon cancer cells. Although the cell viability ratio was reduced, the cells continued dividing after DENSpm treatment (Fig. 1a–c). Thus, p53 profile might have an essential role in the DENSpm (10 μM) triggered cell death induction in colon cancer cells.
The cytotoxic effect of DENSpm in HCT116 p53+/+, SW480, HCT 116 p53−/− colon cancer cells. The effect of DENSpm on cell viability was determined by MTT cell viability assay after DENSpm (0–50 μM) treatments in time-dependent manner (24, 48, 72 h) in a HCT 116 p53+/+, b SW480, c HCT 116 p53−/− colon cancer cells. The columns represent the mean ± SD two independent trial with at least 2 replicates. The effect of DENSpm on cell death induction was determined in each colon cancer cells. 0.5 × 104 cells were treated with 10-µM DENSpm for 72 h at different time points and then cells were stained with trypan blue, viable cells were counted under light microscope. The data shown represent the mean ± SD from two experiments with three replicates. Statistical difference was analyzed using an unpaired t test; *p < 0.05, **p < 0.001, ***p < 0.0001
DENSpm induces the accumulation of subG1 population
To evaluate the potential effect of DENSpm on colon cancer cells, we performed cell cycle analysis following PI staining. Consistent with previous cell proliferation and cell viability results, the subG1 apoptotic population was increased in time-dependent manner (0–72 h) following DENSpm treatment by 3.5, 5.2, 10.9 % percent, respectively, in HCT 116 p53+/+ cells (Fig. 2a). However, DENSpm treatment did not cause a significant induction of the sub G1 population in SW480 colon cancer cells, but this effect was significant after 72 h drug treatment, with an increase of 3.2 % (Fig. 2b). In addition, no significant subG1 population was identified in p53−/− HCT 116 colon cancer cells. For the time-dependent DENSpm treatment, the sub G1 population increased by 2.2, 2.9, and 4.5 %, which was also consistent with the resistant profile of p53 deficient cells for DENSpm treatment (Fig. 2c).
Effects of DENSpm on cell cycle progression. a HCT116 p53+/+, b SW480, c HCT 116 p53−/− colon cancer cells were treated with 10 μM DENSpm for 24, 48, 72 h and then cells were harvested, fixed with ethanol, and stained with PI. The cellular DNA contents were determined by flow cytometer analysis to detect the cell cycle distribution. Results shown were representative of two independent experiments
DENSpm activates PA catabolic machinery
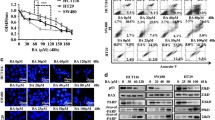
As shown in Fig. 3a, DENSpm treatment for 24 h caused a sharp decrease in the Put, Spd and Spm levels in HCT 116 p53+/+ cells compared to the untreated control cells (*p < 0.01, ***p < 0.0001). This effect was further increased after long-term treatment with DENSpm (for Put levels, 48 h **p = 0.0091, 72 h p < 0.001). To determine the effect of DENSpm on the regulation of the intracellular PA pool, we determined the expression levels of SSAT and PAO (Polyamine oxidase), which are expressed in control cells at very low levels. Exposure of HCT 116 p53+/+ cells to DENSpm upregulated both the SSAT and PAO protein expression levels (Fig. 3a). Consistent with this finding, DENSpm was also involved in PA homeostasis via the regulation of the expression of the PA biosynthetic enzyme ODC (Ornithine decarboxylase) in HCT 116 p53+/+ cells. DENSpm treatment was downregulated expression of both ODC and its activator, AZI (Antizyme Inhibitor), in a time-dependent manner (Fig. 3a). For the PA pool changes in p53 mutant (mt) SW480 cells, we found that DENSpm significantly decreased the PA levels after 48 h. The drug affected only the Put levels within 24 h (*p 0.016), but no significant effect was observed for Spd (p > 0.05) and Spm levels (p > 0.05) compared to the untreated cells. Similar to the HCT 116 p53+/+ colon cancer cells, the lack of basal SSAT expression in SW480 cells was upregulated after DENSpm treatment for 48 h. Exposure of SW480 cells to DENSpm for 48 h also upregulated the PAO expression level as well. This was consistent with the previous data showing a significant decrease in PA levels in SW480 cells. Although 48 h DENSpm treatment downregulated the ODC protein expression, no significant change was observed for AZI protein expression in colon cancer cells (Fig. 3b). We conclude that the mutation R273H in p53 causes a significant decrease in the chemotherapeutic efficiency of DENSpm in SW480 colon cancer cells through its interaction with PA homeostasis. To confirm the role of p53 in DENSpm-modulated PA homeostasis, we determined the PA levels in p53 knockout HCT 116 colon cancer cells after DENSpm treatment. Although exposure of DENSpm for 24 h only decreased the Put levels compared to control cells, longer treatment of DENSpm significantly decreased the PA pool in p53 knockout HCT 116 cells (**p < 0.001) (Fig. 3c). The effectiveness of DENSpm on the PA intracellular pool in p53 knockout HCT 116 cells was also confirmed by SSAT upregulation, particularly after 72 h. In addition, DENSpm did not alter the biosynthetic molecular proteins of the PA signaling cascade in p53−/− HCT 116 cells. To investigate the generation of ROS due to DENSpm-induced PA catabolic enzyme activation, we performed FACS flow analysis following CM-H2DCFDA staining. Based on the FACS flow results, DENSpm-induced ROS generation in the colon cancer cells, regardless of p53 expression, was accelerated in time-dependent manner (Suppl. Figure 1). According to these observations, we concluded that p53 expression in colon cancer cells may be critical for evaluating the therapeutic effect of DENSpm.
DENSpm-induced PA depletion by activating PA catabolic enzymes at time-dependent manner in colon cancer cells. The effect of DENSpm (10 μM) on intracellular PA pool was determined by HPLC analysis (left) and PA metabolic enzymes; SSAT, PAO, ODC, AZI expression profiles following DENSpm treatment were investigated by immunoblotting (right). β-Actin was used as a loading control. HPLC analysis and blotting results shown was representative of two independent experiments
DENSpm activates mitochondria-mediated apoptosis by modulating Bcl-2 family members
Since mitochondria play a critical role in the regulation of apoptotic machinery, we examined mitochondrial membrane potential (Δψm) loss using JC-1 staining after time-dependent DENSpm treatment of HCT 116 p53+/+, SW480 and HCT 116 p53−/− cells. When we compared the time-dependent DENSpm effect on Δψm, a significant increase in Δψm loss was observed in HCT 116 p53+/+ cells (percentage of the depolarized Δψm was 51.9 % in 72 h). However, resistant profile against DENSpm treatment was observed in both SW480 and p53 knockout HCT 116 colon cancer cells only for 48 h (percentage of the depolarized Δψm was 18.8 % in SW480 and 15.1 % in HCT 116 p53−/−). But, prolonged DENSpm treatment (72 h) induced apoptotic induction in each mutant colon cancer cells (percentage of the depolarized Δψm was 42.5 % in SW480 and 38.5 % in HCT 116 p53−/−) (Fig. 4a).
DENSpm-induced apoptosis by activating mitochondrial pathway and modifying Bcl-2 family protein expression. The effect of DENSpm (10 μM) on ∆Ψm was performed by a JC-1 staining by FACS Flow cytometry. After 24, 48, and 72 h DENSpm treatments, HCT 116 p53+/+, SW480, HCT 116 p53−/− colon carcinoma cells were stained with JC-1, and results analyzed by FACS Flow cytometer. The number in the upper side of the curved line each dot plot represents the percentage of cells that has normal polarized mitochondrial membranes (given as percentage of aggregates). The numbers given bottom side of the curved line represent the cells emit only green fluorescence attributable to DENSpm-induced mitochondrial membrane depolarization (given as a percentage of monomers). The effect of DENSpm (10 μM) on b Caspase-3,-9 activation and PARP cleavage and c Bcl-2 family protein expression profiles was determined by immunoblotting in HCT 116 p53+/+ (left), SW480 (middle), HCT 116 p53−/− (right) colon carcinoma cells. β-actin was used as a loading control
To understand the apoptotic potential of DENSpm, we determined caspase activation by identifying the cleavage profiles of caspase-9, and -3 following time-dependent DENSpm treatment of colon cancer cells (Fig. 4b). Whereas DENSpm treatment (10 µM) for 24 h significantly reduced the full length caspase-9 and -3 levels in HCT 116 p53+/+ colon cancer cells, significant caspase-9 and -3 activation was detected after 48 h DENSpm treatment of SW480 cells. Moreover, time-dependent DENSpm induced caspase activation and PARP cleavage was observed only after 72 h of drug treatment in p53 knockout HCT 116 colon cancer cells (Fig. 4b).
To further evaluate the time-dependent effect of DENSpm on mitochondria-mediated apoptosis, we next analyzed the expression profile of Bcl-2 family members in all colon cancer cells. As shown in Fig. 4c, although the anti-apoptotic Bcl-2 and Mcl-1 expression levels were downregulated after 24 h of DENSpm treatment in HCT 116 p53+/+ cells, upregulation of the pro-apoptotic Bcl-2 family members (Puma and Bax) were observed only after 72 h drug exposure. DENSpm treatment for 24 h downregulated the anti-apoptotic Bcl-xL protein and upregulated the pro-apoptotic Bax protein expression in p53 mt SW480 cells. Similar to previous findings, DENSpm did not alter the expression profile of Bcl-2 family members in p53 knockout HCT 116 cells (Fig. 4c). These results indicate that DENSpm induces apoptosis in functional p53-expressing HCT 116 cells; however, a resistance mechanism is evident in p53 mutant or knockout colon cancer cells.
Depletion of PA-induced autophagy and p62 downregulation
To evaluate the effect of DENSpm on the progression of autophagy, HCT 116 (p53+/+), SW480 (p53 mt) and HCT 116 p53−/− colon cancer cells were stained with AO following drug treatment. PA depletion due to activation of catabolic enzymes induced autophagy-mediated vacuole formations after 48 h DENSpm treatment in both HCT 116 p53+/+ (50.1 %), and SW480 cells (41.7 %). However, this effect was not observed following 72 h DENSpm treatment in colon cancer cells due to the induction of apoptosis. Moreover, because untreated p53 null HCT 116 colon cancer cells (92.3 %) have high levels of acidic vacuole formation, HCT 116 p53−/− cells showed a resistant profile to DENSpm (Fig. 5).
Although DENSpm as a PA analogue reduced the intracellular PA pool and activated apoptosis based on the functional p53 status in colon cancer cells, its effect on autophagy has not been fully elucidated yet. To identify the key players in DENSpm-induced cell death, we investigated the autophagy-related gene expressions in colon cancer cells. Although basal Beclin-1 expression was detected in HCT 116 p53+/+ and p53−/− colon cancer cells, there was no significant expression of Beclin-1 determined in SW480 cells. Although Beclin-1 expression was significantly upregulated in time-dependent manner in SW480 cells, downregulation of Beclin-1 expression was observed in HCT 116 p53−/− cells following 48 h DENSpm treatment. Moreover, Beclin-1 expression was slightly upregulated after 48 h DENSpm treatment in HCT 116 p53+/+. Whereas DENSpm downregulated Atg3 expression within 48 h in HCT 116 p53+/+ and SW480 cells, the Atg5 and Atg12 expression levels were downregulated only in HCT 116 p53+/+ cells. Although Atg12 expression was upregulated following 48 h DENSpm treatment in SW480 cells, no significant change was observed for Atg5 expression. Moreover, no significant effect was observed for Atg7 expression level in all colon cancer cells after drug treatment. DENSpm treatment induced cleavage of LC3-I to LC3-II and p62 degradation in HCT 116 p53+/+ and SW480 cells. Although DENSpm modulated the autophagy-signaling mechanism, lack of a functional p53 causes the autophagy inducing effect of DENSpm, which did not have a significant effect on the Atg3, 5, 7 and 12 expression profiles. Conversely, DENSpm-induced LC3 cleavage and p62 degradation were observed in untreated HCT 116 p53−/− colon cancer cells because p53 is essential for the inhibition of autophagy in cancer cells (Fig. 6a–c).
DENSpm-induced autophagy induction colon carcinoma cells in different p53 expression profile. The effect of DENSpm (10 μM) on Beclin-1, LC3, Atg3, Atg5, Atg7, Atg12, p62 gene expression profiles was determined by immunoblotting in a HCT116 p53+/+ , b SW480, c HCT116 p53−/− colon cancer cells following 24, 48, 72 h DENSpm (10 μM) treatments. β-actin was used as a loading control. Representative immunoblotting results were shown from two independent experiments
The role of LC3 in DENSpm-induced autophagy in colon cancer cells
To verify that the autophagy-signaling cascade following DENSpm treatment is dependent on the functional p53 protein, colon cancer cells were transfected with GFP-tagged LC3 and the involvement of LC3 in the DENSpm-induced autophagy regulation was observed in time-dependent manner (Fig. 7a, b). The maximum GFP-LC3 fluorescence due to GFP-LC3 dot formation caused by DENSpm treatment was detected after 48 h in HCT 116 p53+/+ and p53 mt SW480 cells. However, the formation of GFP-LC3 dot ratio was higher in untreated SW480 cells compared to HCT 116 cells, which indicates that p53 mt SW480 cells are more vulnerable to DENSpm-induced autophagy.
Involvement of LC3 in DENSpm-induced autophagy in colon carcinoma cells. Following 24 h transfection of GFP-LC3 plasmid to colon cancer cells, cells were treated with 10-μM DENSpm for 24, 48, 72 h. DENSpm-induced GFP-LC3 formations were examined by FACS flow analysis at FL-1 channel. The graph represents the fluorescence intensity of GFP-LC3 dots due to DENSpm treatment in each cell line. Results are shown as the mean ± SD of two independent experiments. Statistical difference was analyzed using an unpaired t test; *p < 0.05, **p < 0.001
Discussion
DENSpm is a commonly used PA analogue that was investigated in in vitro, in vivo and in clinical studies (Schipper et al. 2000; Wolff et al. 2003) has been reported to induce cell death by activating PA catabolic machinery and generating ROS due to cytotoxic by-products (Ha et al. 1997; Saab et al. 1993). DENSpm inhibits cell proliferation and induces cell cycle arrest in various cancers such as breast (Hegardt et al. 2002), prostate (Schipper et al. 2000), non-small lung (Hahm et al. 2002), melanoma (Chen et al. 2001) and colon cancer (Allen et al. 2007). However, the expression levels of survivin and p53 may alter the therapeutic efficiency of DENSpm in melanoma and colon cancer cells. Although 10-µM DENSpm suppresses cell growth in SK-MEL-28 melanoma cells after 48 h drug treatment, no significant effect was detected for the MALME-3 M and LOX melanoma cell lines (Chen et al. 2003). In addition, p53 null HCT 116 colon cancer cells were resistant to DENSpm in a time- and dose-dependent manner compared to HCT 116 p53 wt colon cancer cells (Choi et al. 2005). In this study, our objective was to determine the role of p53 in therapeutic efficiency of DENSpm in colon cancer cells. We observed that DENSpm decreased cell viability and induced cell death in a dose- and time-dependent manner more effectively in HCT 116 p53 wt colon cancer cells than p53 mutant (SW480) or null (HCT 116 p53−/−) colon cancer cells (Fig. 1a–c). Similar to these findings, we showed that DENSpm (10 µM) treatment reduced the amount of viable cells and induced the cell death which was determined by trypan blue assay. However, the lack of a functional p53 within the cells causes the inhibition of proliferation by DENSpm (Fig. 1a–c). Similarly, although DENSpm induces cell cycle arrest at the G1 phase in MALME-3 M melanoma cells with a functional p53 expression, DENSpm did not induce cell cycle arrest at the G1 phase in SK-MEL-28 p53 mt melanoma cells (Kramer et al. 1999). We determined that DENSpm mediates growth arrest dependent on the p53 expression profile in colon cancer cells. We determined that DENSpm induces an increased subG1 population in HCT 116 wt cells in a time-dependent manner, but there was no significant cell cycle arrest determined in p53 mutant colon cancer cells within 48 h (Fig. 2a–c). Therefore, we concluded that DENSpm-induced cell death and reduction of cell proliferation are dependent on functional p53 in colon cancer cells.
DENSpm induces apoptotic cell death in various cancer cells by depletion of PAs via both upregulation of SSAT and downregulation of ODC. Moreover, it induces PA depletion in MALME-3 M and SK-MEL-28 melanoma cells (Chen et al. 2003) and T24 and J82 bladder cancer cell lines within 24 h (Chang et al. 1993). Moreover, in NCI-H157 non-small lung cancer cells, DENSpm significantly depleted the intracellular PA pool within 24 h (McCloskey et al. 1996). Similarly, time-dependent DENSpm treatment induces intracellular PA pool depletion in various breast cancer cell lines, such as MCF-7, SK-BR-3 and HCC1937 cells. These cells underwent apoptosis following DENSpm treatment due to the sensitivity of the cell cycle at S-phase (Oredsson et al. 2007). Based on these results, we investigated the time-dependent effect of DENSpm on the intracellular PA levels in colon cancer cells with different p53 expression profiles. As shown in Fig. 3a–c, the depletion of the intracellular PA pool was significant in HCT 116 p53 wt colon cancer cells following 24 h drug treatment (Fig. 3a). For the time-dependent effect of DENSpm on mutant p53 expressing SW480 colon cancer cells, we observed a decrease in the intracellular PA pool following 48 h DENSpm treatment (Fig. 3b). However, this depletion was not significant in HCT 116 p53−/− colon cancer cells until 72 h (Fig. 3c). We hypothesized, that p53 mutation, a key cell cycle molecule, may render the therapeutic efficiency of DENSpm in colon cancer cells. In addition, as DENSpm can induce SSAT activity up to 1,000-fold, SSAT enzyme activity was postulated to be responsible for the anti-proliferative effect in cancer cells (Porter et al. 1991). Chinese hamster ovary cells (CHO) were resistant to DENSpm because of a point mutation in the SSAT gene (McCloskey and Pegg 2000). Moreover, silencing of SSAT prevented the DENSpm-induced apoptosis and caspase activation in SK-MEL-28 melanoma cells (Chen et al. 2003). Therefore, we investigated the time-dependent effect of DENSpm on the PA metabolic enzymes expression profiles in colon cancer cells with different functional p53 protein (Fig. 3a–c). Although DENSpm induced SSAT expression after 24 h treatment in HCT 116 p53+/+ colon cancer cells, significant induction of SSAT was determined only after 48 h in SW480 p53 mt and 72 h in HCT 116 p53−/− colon cancer cells. When we compared drug-specific responses of colon cancer cells against DENSpm treatment, p53 mt and/or p53 absence caused late transcriptional response against DENSpm treatment compared to p53 wt cells. Thus, we assumed that functional p53 is related to SSAT expression and is essential in colon cancer cells for understanding the therapeutic effect of DENSpm.
p53 is a major DNA repair molecule and tumor suppressor that is frequently mutated in human cancers and is activated by apoptotic stimuli. Inactivation of p53 causes resistance to chemotherapeutic drugs in cancer cells (Kim et al. 2009). Consequently, in this study we investigated the role of p53 in DENSpm-induced mitochondria-mediated apoptosis in colon cancer cells. We found that time-dependent exposure of wt p53 expressing HCT 116 cells to DENSpm-induced Δψm loss, caspase-3, -9, -7 activation and PARP cleavage. Although, DENSpm treatment for 48 h induced mitochondria-mediated apoptotic cell death in HCT 116 cells, the presence of mt p53 protein prevented the DENSpm-induced apoptosis (Fig. 4a, b). It was reported that DENSpm could induce apoptotic cell death in a p53-dependent manner in breast cancer cells (Hegardt et al. 2002). However, there are contradictory results which showed that DENSpm induces apoptotic cell death without affecting cell cycle arrest in p53 mt SK-MEL-28 melanoma cells through triggering cytochrome c release and caspase activation in time-dependent manner (Chen et al. 2001).
Apoptotic chemotherapeutic agents in clinical use for colon and cervical cancer such as oxalaplatin, cisplatin and 5-FU were shown to cause drug resistance and cancer recurrence (Li et al. 2009; Xu et al. 2012). Therefore, investigating different drug resistance mechanisms, such as the induction of autophagy, has gained importance. Despite the well-known role of DENSpm in apoptosis, the autophagic response of different cancer cells against this PA analogue is not known. Therefore, in this study our objective was to determine the autophagic regulation by DENSpm in colon carcinoma cells with different p53 expression profiles. We found that Beclin-1 was upregulated and LC3-II formation and p62 degradation were induced after 24 h DENSpm treatment in HCT 116 p53 wt colon cancer cells. We found that longer exposure of cells to DENSpm, which exhibited severe apoptotic induction, the autophagy markers Atg5 and Atg12 were downregulated compared to untreated control cells (Fig. 6a). DENSpm induced autophagy in p53 mutant SW480 colon cancer cells via upregulating Beclin-1 and Atg12 expression levels and increasing p62 degradation occurred after 48 h DENSpm treatment (Fig. 6b). However, resistance to DENSpm was obvious due to the induction of autophagy through basal Beclin-1 expression, LC3-II formation and p62 degradation in untreated HCT 116 p53−/− colon cancer cells (Fig. 6c). Similarly, time-dependent DENSpm induced autophagy regulation in HCT 116 colon cancer cells as determined by GFP-LC3 dot formation and AO uptake (Figs. 5, 6, 7). This different regulation of DENSpm-induced autophagy/apoptosis may be due to p53 expression profile. Recently, p53 was reported as a key molecule both in apoptosis and autophagy because of to its localization within the cell (Morselli et al. 2008b). Although nuclear localization of p53 acts as a transcription factor and induces Puma, Noxa, and Bax expressions, it can also induce autophagy through the regulation of the autophagy-related gene expressions. Moreover, it was shown that cytoplasmic p53 may inhibit autophagy in HCT 116 colon cancer cells (Morselli et al. 2008a).
Although DENSpm induces apoptotic cell death in both p53 wt and mutant colon cancer cells, functional p53 activates the apoptotic machinery after DENSpm treatment in HCT 116 colon cancer cells, which caused significant PA depletion and activation of catabolic enzymes. However, autophagy regulation is activated prior to apoptosis in p53 mutant colon cancer cells, and this activation may delay DENSpm-induced apoptosis in SW480 colon cancer cells. This is the first report showing that DENSpm induces autophagy in addition to its apoptotic effect in colon cancer cells. Furthermore, p53 is an important key molecule in autophagy/apoptosis caused by DENSpm treatment. To increase the therapeutic effect of DENSpm in colon cancer cells, autophagy inhibition may be an essential target to overcome the resistance of colon cancer to DENSpm.
Abbreviations
- AO:
-
Acridine orange,
- AZI:
-
Antizyme inhibitor
- BH:
-
Bcl-2 homology
- CHENSpm:
-
N1-ethyl-N11-((cycloheptyl)methyl)-4,8-diazaundecane
- CHO:
-
Chinese hamster ovary cell
- DENSpm:
-
N1, N11-diethylnorspermine
- DMSO:
-
Dimethylsulfoxide
- MTT:
-
3-4,5-Dimethyl-2-thiazolyl-2,5-diphenyl-2H-tetrazolium bromide
- JC-1:
-
5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethyl benzimidazol carbocynanne iodide
- ODC:
-
Ornithine decarboxylase
- PA:
-
Polyamine
- PAO:
-
Polyamine oxidase
- PBS:
-
Phosphate-buffered saline
- PI:
-
Propidium iodide
- Put:
-
Putrescine
- PVDF:
-
Polyvinyldifluoride
- SDS-PAGE:
-
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
- Spd:
-
Spermidine
- Spm:
-
Spermine
- SSAT:
-
Spermidine/spermine N1-acetyltransferase
- TBS:
-
Tris-buffered saline
- Δψm:
-
Mitochondrial membrane potential
References
Allen WL et al (2007) The role of spermidine/spermine N1-acetyltransferase in determining response to chemotherapeutic agents in colorectal cancer cells. Mol Cancer Ther 6:128–137. doi:10.1158/1535-7163.MCT-06-0303
Chang BK, Liang Y, Miller DW, Bergeron RJ, Porter CW, Wang G (1993) Effects of diethyl spermine analogues in human bladder cancer cell lines in culture. J Urol 150:1293–1297
Chen Y, Kramer DL, Diegelman P, Vujcic S, Porter CW (2001) Apoptotic signaling in polyamine analogue-treated SK-MEL-28 human melanoma cells. Cancer Res 61:6437–6444
Chen Y, Kramer DL, Li F, Porter CW (2003) Loss of inhibitor of apoptosis proteins as a determinant of polyamine analog-induced apoptosis in human melanoma cells. Oncogene 22:4964–4972. doi:10.1038/sj.onc.1206725
Choi W et al (2005) Combination of 5-fluorouracil and N1, N11-diethylnorspermine markedly activates spermidine/spermine N1-acetyltransferase expression, depletes polyamines, and synergistically induces apoptosis in colon carcinoma cells. J Biol Chem 280:3295–3304. doi:10.1074/jbc.M409930200
Cohen SS (1978) What do the polyamines do? Nature 274:209–210
Crighton D et al (2006) DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126:121–134. doi:10.1016/j.cell.2006.05.034
Ha HC, Woster PM, Yager JD, Casero RA Jr (1997) The role of polyamine catabolism in polyamine analogue-induced programmed cell death. Proc Natl Acad Sci USA 94:11557–11562
Hahm HA, Ettinger DS, Bowling K, Hoker B, Chen TL, Zabelina Y, Casero RA Jr (2002) Phase I study of N(1), N(11)-diethylnorspermine in patients with non-small cell lung cancer clinical cancer research : an official journal of the American Association for. Cancer Res 8:684–690
Hector S et al (2004) Polyamine catabolism in platinum drug action: interactions between oxaliplatin and the polyamine analogue N1, N11-diethylnorspermine at the level of spermidine/spermine N1-acetyltransferase. Mol Cancer Ther 3:813–822
Hegardt C, Johannsson OT, Oredsson SM (2002) Rapid caspase-dependent cell death in cultured human breast cancer cells induced by the polyamine analogue N(1), N(11)-diethylnorspermine. Eur J Biochem/FEBS 269:1033–1039
Igarashi K, Kashiwagi K (2000) Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun 271:559–564. doi:10.1006/bbrc.2000.2601
Kim E, Giese A, Deppert W (2009) Wild-type p53 in cancer cells: when a guardian turns into a blackguard. Biochem Pharmacol 77:11–20. doi:10.1016/j.bcp.2008.08.030
Kramer DL et al (1999) Polyamine analogue induction of the p53-p21WAF1/CIP1-Rb pathway and G1 arrest in human melanoma cells. Cancer Res 59:1278–1286
Li J, Hou N, Faried A, Tsutsumi S, Takeuchi T, Kuwano H (2009) Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann Surg Oncol 16:761–771. doi:10.1245/s10434-008-0260-0
McCloskey DE, Pegg AE (2000) Altered spermidine/spermine N1-acetyltransferase activity as a mechanism of cellular resistance to bis(ethyl)polyamine analogues. J Biol Chem 275:28708–28714. doi:10.1074/jbc.M004120200
McCloskey DE, Yang J, Woster PM, Davidson NE, Casero RA Jr (1996) Polyamine analogue induction of programmed cell death in human lung tumor cells. Clin Cancer Res 2:441–446
Mizushima N (2009) Physiological functions of autophagy. Curr Top Microbiol Immunol 335:71–84. doi:10.1007/978-3-642-00302-8_3
Morselli E, Galluzzi L, Kroemer G (2008a) Mechanisms of p53-mediated mitochondrial membrane permeabilization. Cell Res 18:708–710. doi:10.1038/cr.2008.77
Morselli E et al (2008b) Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle 7:3056–3061
Oredsson SM, Alm K, Dahlberg E, Holst CM, Johansson VM, Myhre L, Soderstjerna E (2007) Inhibition of cell proliferation and induction of apoptosis by N(1), N(11)-diethylnorspermine-induced polyamine pool reduction. Biochem Soc Trans 35:405–409. doi:10.1042/BST0350405
Pattingre S, Levine B (2006) Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res 66:2885–2888
Pegg AE (1988) Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res 48:759–774
Porter CW, Ganis B, Libby PR, Bergeron RJ (1991) Correlations between polyamine analogue-induced increases in spermidine/spermine N1-acetyltransferase activity, polyamine pool depletion, and growth inhibition in human melanoma cell lines. Cancer Res 51:3715–3720
Saab NH, West EE, Bieszk NC, Preuss CV, Mank AR, Casero RA Jr, Woster PM (1993) Synthesis and evaluation of unsymmetrically substituted polyamine analogues as modulators of human spermidine/spermine-N1-acetyltransferase (SSAT) and as potential antitumor agents. J Med Chem 36:2998–3004
Schipper RG, Deli G, Deloyer P, Lange WP, Schalken JA, Verhofstad AA (2000) Antitumor activity of the polyamine analog N(1), N(11)-diethylnorspermine against human prostate carcinoma cells. Prostate 44:313–321
Wolff AC et al (2003) A Phase II study of the polyamine analog N1, N11-diethylnorspermine (DENSpm) daily for 5 days every 21 days in patients with previously treated metastatic breast cancer. Clin Cancer Res 9:5922–5928
Xu Y et al (2012) Inhibition of autophagy enhances cisplatin cytotoxicity through endoplasmic reticulum stress in human cervical cancer cells. Cancer Lett 314:232–243. doi:10.1016/j.canlet.2011.09.034
Yang Z, Klionsky DJ (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335:1–32. doi:10.1007/978-3-642-00302-8_1
Zagaja GP, Shrivastav M, Fleig MJ, Marton LJ, Rinker-Schaeffer CW, Dolan ME (1998) Effects of polyamine analogues on prostatic adenocarcinoma cells in vitro and in vivo. Cancer Chemother Pharmacol 41:505–512
Acknowledgments
This work was supported by the grant from TUBITAK (TBAG-212T227) and funded in part by Istanbul Kultur University Scientific Projects Support Center.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl 1. DENSpm-induced ROS generation. ROS generation following PA catabolic enzyme activation was determined by CM-H2DCFDA staining in A) HCT 116 p53+/+, B) SW480 and C) HCT 116 p53−/− colon cancer cells. β-actin was used as a loading control. Representative immunoblotting results were shown from two independent experiments.
Rights and permissions
About this article
Cite this article
Çoker-Gürkan, A., Arisan, E.D., Obakan, P. et al. Lack of functional p53 renders DENSpm-induced autophagy and apoptosis in time dependent manner in colon cancer cells. Amino Acids 47, 87–100 (2015). https://doi.org/10.1007/s00726-014-1851-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-014-1851-7