Abstract
Lycium ruthenicum Murr. is an important ecological and economic species in the Qaidam Basin of Qinghai–Tibet Plateau. Its black fruits (BF) are rich in anthocyanins, which have health-promoting properties for humans and thus provide nutritional benefits for this plant. Although the fruit quality of natural white fruit (WF) is affected by the disappearance of pigmentation in phenotypes, this phenomenon provides an opportunity to unravel the complex color metabolic networks. In this study, anthocyanin profiling confirmed that WF was formed due to anthocyanin loss. Transcriptome analysis of BF and WF revealed 101,466 unigenes, 261 of which were identified as the putative homologs of color-related genes in other species. Genes encoding the enzymes involved in flavonoid biosynthesis were also identified systematically. The structural gene expression levels of chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid 3′5′-hydroxylase (F3′5′H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and anthocyanidin 3-O-glucosyltransferase (UFGT) were highly similar and significantly positively correlated with anthocyanin accumulation rate in BF. In particular, F3′5′H, UFGT, ANS, and DFR expression levels in BF were 2391, 119, 96, and 85 times higher than those in WF at S3 (35 days after anthesis), respectively. This result strongly suggests that the low expression of these genes in WF is responsible for the anthocyanin loss. Meanwhile, the expression patterns of the anthocyanin regulatory genes were also investigated by qRT-PCR. Mass sequencing data were obtained and annotated by deep sequencing and provided a platform for future function and molecular biological research on L. ruthenicum Murr.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lycium ruthenicum Murr. is a rare shrub plant belonging to the Solanaceae family. This wild-growing species is ideal for preventing soil desertification and alleviating soil salinity–alkalinity because of its stress, drought, and salt tolerance and resistance to diseases and pest insects (Zheng et al. 2011). Owing to the abundance of anthocyanins in the black fruits (BF) of this plant (Zheng et al. 2011; Peng et al. 2012; Altintas et al. 2006; Zeng et al. 2014a), it has been used as a traditional medicine in China and exhibits curative effects on heart disease, abnormal menstruation, and menopause (Potterat 2010; Zheng et al. 2011; Zeng et al. 2014a). Therefore, this plant is an important ecological and economic species in the Qaidam Basin of Qinghai–Tibet Plateau.
Owing to the high altitude, cold, dry climate, strong sunshine, and saliferous soil in Qinghai–Tibet Plateau, the anthocyanins accumulate in the fruit, resulting in the black pigments. The anthocyanin prevents UV-B damage and subsequent cell death by protecting the DNA from dimerization and breakage for survival in Qinghai Province (Zheng et al. 2011). Interestingly, white fruit (WF) is also found in nature at the resource inventory of the Qinghai Province in 2013–2014, and subsequent research (Zhang et al. 2018) indicates that the WF is a mutant of L. ruthenicum Murr. The disappearance of the anthocyanins in phenotypes may reduce the plant’s ability to adapt to the environment of Qaidam Basin and greatly influence the berry quality. Natural phenotypic variation provides an opportunity to unravel the role of anthocyanin genes that lead to the extreme colors of L. ruthenicum Murr.
As the examples of flavonoids, anthocyanins are among the most studied and understood compounds, and their metabolic pathway in plants has been extensively described (Grotewold 2006; Tanaka et al. 2008). However, the underlying molecular mechanisms that control anthocyanin catabolism in different plant species remain unclear. In particular, loss-of-color adaptations are relatively unrestrained in different species because they can be achieved in many ways (Lou et al. 2014). Blocking an early gene in anthocyanin biosynthetic pathway could be more efficient, and thus CHS mutation is the most common method to induce loss-of-color in flowers or fruits (Clark and Verwoerd 2011), followed by the absence of more than one enzyme in anthocyanin biosynthetic pathway (Bogs et al. 2007; Clark and Verwoerd 2011). Recent research showed that the lack of color phenotype can be determined by regulating the branching point of anthocyanin biosynthesis (Lou et al. 2014). Altering the substrate competition between flavonol synthase (FLS) and dihydroflavonol 4-reductase (DFR) may eliminate the blue pigmentation in grape hyacinth (Lou et al. 2014). Introducing apple ANR genes into tobacco inhibits the expression of CHI and DFR genes in flowers, finally leading to anthocyanin loss (Han et al. 2012). FcMYB1 determines the discolored phenotype of white Chilean strawberry fruit by regulating the branching point of anthocyanin/proanthocyanidin biosynthesis (Salvatierra et al. 2013).
In this study, RNA-seq for L. ruthenicum Murr. and its white variant was performed using Illumina technology to comprehensively analyze and determine the mechanism for pigmentation loss in its fruits. Bioinformatics analysis revealed the sets of up- and downregulated genes from BFs and WFs and the candidate genes targeting pigmentation loss were examined. The annotated transcriptome sequences provide a valuable genomic resource and platform to further understand the molecular basis of fruit color variations in L. ruthenicum Murr. and other members of Solanaceae family.
Materials and methods
Plant material
In September 2015, BF and WF (healthy fruits under natural light condition) L. ruthenicum Murr. samples and its white form used in this work were collected from Zongjia Town, Dulan County, Qinghai Province, P. R. China. The samples were harvested during the three fruit developmental stages (S1–S3) based on fruit epidermis pigmentation and are shown in Fig. 1a with the following characteristics: unripe green fruit before color breaker (BS1, WS1, 5 days after anthesis); the light-color stage, in which the BF appears light aubergine and the WF appears yellowish white (BS2, WS2, 15 days after anthesis); and the stage of BFs and WFs (BS3, WS3, 35 days after anthesis). The fruits were manually collected with scissors while avoiding any mechanical damages. For each developmental stage, three replicate groups were prepared for physiological and biochemical determination. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C for pending measurement.
Coloration phenotype and anthocyanin content in developing L. ruthenicum Murr. fruits. a Coloration phenotype of fruits at three developmental stages, bar = 5 mm. b Anthocyanin contents in fruits at three developmental stages. Total anthocyanin content in ripening fruits indicated as the mean ± SD of three replicates
Anthocyanin determination
Anthocyanin in L. ruthenicum Murr. fruits was extracted and determined as described previously (Zeng et al. 2014a; Qi et al. 2019). The petunidin derivatives accounted for 95% of the total anthocyanins in fresh BF that is derived from the delphinidin branch and mediated by F3′5′H activity, and the remaining 5% are also derivatives of delphinidin (Zheng et al. 2011). Therefore, delphinidin chloride (ChromaDex, Santa Ana, CA, USA) was used as standard for quantification. Mean values and standard deviations (SDs) were obtained from three biological replicates.
RNA extraction
Total RNA was extracted from tissues using plant RNA purification reagent according the manufacturer’s instructions (Invitrogen). Genomic DNA was removed using DNase I (TaKara), and RNA quality was determined by 2100 Bioanalyzer (Agilent) and quantified using ND-2000 (NanoDrop Technologies, USA). Only high-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 10 μg) was used to construct the sequencing library.
Library preparation and Illumina HiSeq 4000 sequencing
RNA-seq transcriptome library was prepared following the instruction from TruSeq™ RNA sample preparation kit from Illumina (San Diego, CA) using 5 μg of total RNA. Messenger RNA was isolated according to polyA selection by oligo(dT) beads and then fragmented by using a fragmentation buffer. Double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina) and then subjected to end-repair, phosphorylation, and “A” base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 200–300 bp on 2% low-range ultra-agarose, followed by PCR amplification using Phusion DNA Polymerase (NEB) for 15 PCR cycles. After quantification by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq 4000 (2 × 150 bp read length).
De novo assembly and annotation
Raw data were first processed through in-house Perl scripts. Clean data/reads were obtained by removing adapter- and ploy-N-containing and low-quality reads from raw data. Q20, Q30, GC-content, and sequence duplication of clean data were calculated. The obtained clean data were used for assembly with Trinity (https://github.com/trinityrnaseq/trinityrnaseq/wiki). Gene function was annotated based on the following databases: NR (NCBI non-redundant protein sequences), Pfam (homologous protein family), KOG/COG/eggNOG (clusters of orthologous groups of proteins), Swiss-Prot (manually annotated and reviewed protein sequence database), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO).
Differential expression and functional enrichment analyses
Differential expression analysis between the two groups was performed using DESeq (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html). DESeq provides statistical routines for determining differential expression in digital gene expression data by using a model based on the negative binomial distribution. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling false discovery rate (FDR). The screening criteria were FDR < 0.01 and fold change ≥ 2. In addition, GO functional enrichment and KEGG pathway analyses were performed to distinguish the differentially expressed genes (DEGs) that were significantly enriched in GO terms and metabolic pathways by using the topGO R packages (http://www.bioconductor.org/packages/release/bioc/html/topGO.html) based on Kolmogorov–Smirnov test and KOBAS (http://kobas.cbi.pku.edu.cn/help.do).
Gene validation and expression analysis
The total RNA from the fruits was extracted as described above, and cDNA was synthesized from the total RNA (1 μg) using PrimeScript™ RT Master Mix Kit to reach 20 μl total volume according to the manufacturer’s instructions (TaKaRa). Transcript levels were analyzed by quantitative real-time PCR (qRT-PCR) using the SYBR® Premix Ex Taq™ II (TaKaRa) and a CFX Connect Real-time PCR Cycler (Bio-Rad Laboratories, USA) according to the manufacturers’ instructions. All biological replicates were analyzed in triplicate. EF1α gene was used as the internal control for normalizing gene expression (Zeng et al. 2014b), and the primers were designed using the Primer Premier software (Supplementary Table S1). The relative expression levels of the target genes were calculated using 2-△△CT.
Results
Anthocyanin levels in the fruit of L. ruthenicum Murr at three developmental stages
The anthocyanin content in fruits at different development stages was determined to provide a basis for the lack of color phenotype in WF. The fruits were sampled at three developmental stages (S1–S3) according to BF phenotype (Fig. 1a), which represents visually increasing fruit pigmentation and presumably increasing anthocyanin content. The results showed that the anthocyanin content remained at an extremely low level in both fruits at S1. For BF, the amount increased from S2 and reached a noticeable degree at stage S3. However, it did not show any changes in WF at S2–S3 (Fig. 1b) and still remained at an extremely low level. Therefore, the change in fruit color phenotypes in L. ruthenicum Murr. is mainly caused by the anthocyanin content, which starts to diminish at the early developmental stage in WF.
RNA-seq and sequence assembly
High-throughput sequencing was conducted to generate 18 libraries from two samples (BF and WF) at three fruit developmental stages with three biological replicates to understand the molecular basis of fruit color polymorphism in L. ruthenicum Murr. (see “Materials and methods” for detailed sample information). A total of 183.00 Gb clean data were generated from the 18 libraries, with the pair-end reads in clean data of each sample of not less than 20.65 million, Q30 of each sample ≥ 92.40%. All sequencing data were deposited in NCBI Short Read Archive under BioProject ID PRJNA638483 (accession numbers SRR12149104, SRR12149105, SRR12149106, SRR12149107, SRR12149108, SRR12149109, SRR12149110, SRR12149111, SRR12149112, SRR12149113, SRR12149114, SRR12149115, SRR12149116, SRR12149117, SRR12149118, SRR12149119, SRR12149120, SRR12149121, respectively). The generated high-quality pair-end reads of de novo assembly, which was performed by using Trinity software according to standard parameters, were retrieved after trimming. The assembly produced 201,581 transcripts with a N50 of 1826 bp, and the unigene dataset included 101,466 sequences with a N50 of 1492 bp. The distribution of length of transcripts and unigenes which were used for annotation analysis is shown in Fig. S1a, b. The public databases including COG, GO, KEGG, KOG, Pfam, Swiss-Prot, eggNOG, and NR were searched for homologous sequences. A total of 54,986 unigenes were found in at least one of these databases (Table 1), and detailed annotation information was listed (Supplementary Table S2).
Differentially expressed genes during fruit development
The differentially expressed unigenes between BF and WF libraries were first analyzed to investigate differences in unigene expression in BF and WF during development. For the unigene comparison in BF, that is, BS1 versus BS2, BS2 versus BS3, and BS1 versus BS3, 2260, 5465, and 8212 unigenes were differentially expressed, respectively; 1342, 2543, and 4126 were upregulated, respectively; and 918, 2922, and 4086 were downregulated, respectively. For the unigene comparison in WF, that is, WS1 versus WS2, WS2 versus WS3, and WS1 versus WS3, 2175, 3585, and 7172 unigenes were differentially expressed, respectively; 1072, 1231, and 2813 were upregulated, respectively; and 1103, 2354, and 4359 were downregulated, respectively. For the DEG comparison between WF and BF, that is, WS1 versus BS1, WS2 versus BS2, and WS3 versus BS3, 3185, 3962, and 4910 were unigenes, respectively; 1967, 2952, and 3717 were upregulated, respectively; and 1218, 1010, and 1193 were downregulated, respectively (Table 2).
KEGG pathway enrichment analysis of DEGs during fruit development
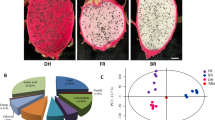
The DEGs were characterized by KEGG pathway enrichment analysis to explore the relevant biological functions and depict the functional differences among the developmental stages of BF and WF in L. ruthenicum Murr. (Fig. 2, and see for full list of KEGG pathway in Supplementary Table S3). Comparisons of these KEGG pathway lists in BF, that is, BS1 versus BS2, BS2 versus BS3, and BS1 versus BS3, revealed five identical pathways, namely, cutin, suberine, and wax biosynthesis; carotenoid biosynthesis; plant hormone signal transduction; phenylpropanoid biosynthesis; and starch and sucrose metabolism (Supplementary Table S3). The pathway of flavonoid biosynthesis appeared in BS2 versus BS3 and BS1 versus BS3 but not in BS1 versus BS2, suggesting no remarkable difference in the gene expression on the pigment pathway at stages 1 and 2 (Supplementary Table S3). This result was in agreement with those on morphological changes and gene expression profiling. In WF, 11 similar pathways were found in WS1 versus WS2, WS2 versus WS3, and WS1 versus WS3. Flavonoid biosynthesis was significantly different in all three groups, and this finding was not consistent with morphological observations (Supplementary Table S3). Comparative analysis for BF and WF revealed that WS1 versus BS1 differed from WS2 versus BS2 and WS3 versus BS3 in terms of KEGG pathway composition, and only one identical difference pathway, namely, flavonoid biosynthesis, was found among the three comparisons. Meanwhile, anthocyanin biosynthesis was only identified in WS1 versus BS1, suggesting that the differences in the expression of anthocyanin synthetic genes in BF and WF were initiated early in the development (Fig. 2, Supplementary Table S3).
Top 20 enriched KEGG pathways of DEGs between BF and WF at different developmental stages. a Scatterplot of enriched KEGG pathway between BF and WF at stage 1. b Scatterplot of enriched KEGG pathway between BF and WF at stage 2. c Scatterplot of enriched KEGG pathway between BF and WF at stage 3. Y-axis on the left represents KEGG pathways, and X-axis indicates the enrichment factor. Pathways with q-values less than 0.05 are significantly enriched. The size of the spot reflects the number of DEGs, and the color of the spot corresponds to different q-value ranges
Defining DEGs related to color development during the developmental stages of BF and WF
The genes involved in the secondary metabolic pathways of pigmentation (flavonoid, anthocyanin, flavones, flavonol, and flavanone biosynthesis) were analyzed using 18 libraries. A total of 261 unigenes were assigned to these pathways. Digital expression analysis identified that all these unigenes were differentially expressed between BF and WF (Supplementary Table S4) and thus were selected for further study.
DEGs between samples were identified to investigate molecular differences between BF and WF under various developmental stages. Expression levels were compared among stages to identify significant DEGs. Table 3 revealed 20, 65, and 83 DEGs in BS1 versus BS2, BS2 versus BS3, and BS1 versus BS3, respectively, and 39, 40, and 79 DEGs in WS1 versus WS2, WS2 versus WS3, and WS1 versus WS3 (Table 3), respectively. The numbers of DEGs in BS2 versus BS3 and BS1 versus BS3 were greater than that in BS1 versus BS2, indicating the involvement of complex developmental events at stage 2.
Table 3 shows that in the comparisons of BS1 versus BS2, BS2 versus BS3, BS1 versus BS3, WS1 versus WS2, WS2 versus WS3, and WS1 versus WS3, 11, 36, 39, 26, 22, and 46 were upregulated, respectively, and 9, 29, 44, 13, 18, and 33 were downregulated, respectively.
Comparison of the transcriptional profiles of genes involved in anthocyanin metabolism between BF and WF
The color distinction between the BF and WF of L. ruthenicum Murr. is caused by the difference of anthocyanin content (Fig. 1). The DEGs between BF and WF were identified to compare their molecular differences under various developmental stages. In comparisons of WS1 versus BS1, WS2 versus BS2, and WS3 versus BS3, 35, 41, and 41 were DEGs, respectively, 28, 26, and 29 were upregulated, respectively; and 7, 15, and 12 were downregulated, respectively. The number of upregulated genes was remarkably higher than that of downregulated ones at all three fruit developmental stages. This phenomenon indicated that anthocyanin loss is due to the low transcript abundance of many genes and was initiated during the early development of the fruit. This result was in agreement with the markedly phenotypic changes in fruit color at S2 and S3 (Fig. 1a).
Genes related to anthocyanin biosynthesis
A total of 261 DEGs related to fruit pigmentation were searched based on standard gene names and synonyms in the combined functional annotations and analyzed to identify the key ones for black-to-white transition. The structural genes (55), including CHS, CHI, flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), F3′5′H, DFR, ANS, UFGT, anthocyanin 5-aromatic acyltransferase (5AT), anthocyanin 5-O-glucoside-6‴-O-malonyltransferase (5MaT1), flavonol synthase (FLS), and anthocyanidin reductase (ANR), and regulatory genes or transcription factors (TFs) (10) participating in each step of the flavonoid biosynthesis, such as MYB, bZIP, C2H2, and HD-Zip, were subsequently identified from the 261 DEG database and studied in detail. A heat map representing the transcript levels for these candidate transcripts in BF and WF at different stages was generated (Fig. 3).
Hierarchical clustering of DEGs in BF and WF of L. ruthenicum Murr.. Each column (WS1 1, WS1 2, WS1 3, WS2 1, WS2 2, WS2 3, WS3 1, WS3 2, WS3 3, BS1 1, BS1 2, BS1 3, BS2 1, BS2 2, BS2 3, BS3 1, BS3 2, and BS3 3) represents the expression value of each RNA-Seq, and each row represents a differentially expressed gene. WS1 1, WS1 2, WS1 3, WS2 1, WS2 2, WS2 3, WS3 1, WS3 2, and WS3 3: WF at three developmental stages (S1–S3, every stage included three biological replicates), respectively; BS1 1, BS1 2, BS1 3, BS2 1, BS2 2, BS2 3, BS3 1, BS3 2, and BS3 3: BF at three developmental stages (S1–S3, every stage included three biological replicates), respectively. The levels are color coded: red indicates high expression levels, and blue indicates low expression levels
Expression profiles of anthocyanin biosynthesis structural genes in BF and WF
Among the six CHS genes identified in BF, c105048, c89559, and c90263 had the lowest expression at S1 and highest expression at S3, c61114 expression exhibited poor repeatability, and the remaining genes showed relatively constant expression during coloration. Two distinct expression patterns were noticed among the three CHI genes. The transcription levels of c103053 and c101670 were low in S1 but high in S3. Opposite expression pattern was observed for c83433. Three F3H genes (c87407, c123966, and c185382) were identified in the fruit transcriptome of L. ruthenicum Murr. c87407 upregulation was evident during coloration, and c123966 and c185382 expression levels were low with minimal changes during development. In the five F3′H genes, c98294, c19510, and c73739 had similar expression patterns and were downregulated with maturity, whereas c86844 showed the opposite expression pattern. c54636 was lowly expressed at S2 but highly expressed at S1 and S3. Three genes (c102345, c138562, and c27994) corresponding for F3′5′H displayed different expression patterns, except for c138562 with poor repeatability. Highest expression of c102345 was detected in S3 and that of c27994 was detected in S1. Five of the DEGs, namely, c92376, c104645, c104723, c107855, and c40236, encoded the predicted DFR. c92376 and c40236 expression increased during coloration, and c104645 and c104723 showed the opposite expression pattern. Among the four genes encoding ANS, c70865 was upregulated from the green to the black stage, c83989 was highly expressed at the green stage, and the remaining genes showed relatively constant expression during coloration. The increasing expression of the 12 genes was correlated with anthocyanin accumulation in five transcripts (c87418, c103977, c85443, c90215, and c99939), whereas the opposite expression pattern was observed for c84984. Moreover, the transcription levels of c103688, c103507, and c38776 were the lowest in the transition period of fruit color (S2). The other three UFGTs (c101538, c95396, and c38776) showed relatively constant expression during coloration. Three genes (c91064, c88939, and c27976) corresponding for 5AT displayed the same expression pattern: the highest expression was detected at the early stage of fruit development, followed by a decrease in levels. Among the 5MaT1 genes, c37514, c74476, c39903, and c82232 showed relatively high and stable expression throughout the development period, and c60991 was not included due to its poor repeatability. The gene-encoding enzymes involved in other subgroups of flavonoid end products were also investigated. For example, three genes specifically participating in proanthocyanidin biosynthesis and encoding anthocyanidin reductase (ANR, c97618, c105446, and c79009) were identified in the database of L. ruthenicum Murr. c97618 and c105446 were downregulated during fruit development, and c79009 showed relatively constant expression at S1–S2 but was upregulated sharply during ripening. Three flavonol synthase genes, namely, c63178, c106360, and c89254, showed the opposite pattern. The expression of the latter was the lowest at S2, whereas the expression of the former was the highest at S2.
The expression patterns of 55 structural genes in WF were also analyzed (Fig. 3). c89559 (CHS), c81030 (CHS), c90263 (CHS), c87407 (F3H), c86844 (F3′H), c98294 (F3′H), c54636 (F3′H), c83989 (ANS), c87418 (UFGT), c103688 (UFGT), c103507 (UFGT), c101538 (UFGT), c85443 (UFGT), c90215 (UFGT), c93992 (UFGT), c91064 (5AT), c74476 (5MaT1), c39903 (5MaT1), c63178 (FLS), and c106360 (FLS) had similar expression pattern of gradual increase, whereas the opposite pattern was observed for c100418 (CHS), c35393 (ANS), c88939 (5AT), c27976 (5AT), and c82232 (5MaT1) with downregulated expression during fruit ripening. c107855 (DFR) showed high expression in S1, and the remaining genes belonging to 12 families maintained a relatively low expression throughout the development period.
Comparison of the expression profiles of anthocyanin biosynthesis structural genes between BF and WF
Comparison of the expression patterns of 55 structural genes in BF and WF revealed that 26 genes showed different patterns and were arranged into four groups (Fig. 3) according to the expression pattern in WF. Group 1 was the largest group containing 14 genes (54%), which were all underexpressed during the whole growth period of WF. In BF, nine genes (c105048 (CHS), c103053 (CHI), c101670 (CHI), c102345 (F3′5′H), c92376 (DFR), c40236(DFR), c70865 (ANS), c103977 (UFGT), and c79009 (ANR)) were upregulated, four genes (c104723 (DFR), c84984 (UFGT), c97618 (ANR), and c105446 (ANR)) were downregulated, and one gene (c37514 (5MaT1)) was highly expressed throughout the development period. Nine of the genes (34%) in group 2 were upregulated in WF along with the fruit ripening. In BF, three genes (c89559 (CHS), c86844 (F3′H), and c87418 (UFGT)) were upregulated. Although showing the same trend, their expression was higher than that in WF. In addition, two genes (c101538 (UFGT) and c74476 (5MaT1)) maintained high expression during development, another two (c54636 (F3′H) and c103688 (UFGT)) were downregulated from green-to-light color stage but upregulated from light-to-black color stage, and the last two (c98294 (F3′H) and c91064 (5AT)) were downregulated. Group 3 contained only two genes (8%), both of which were downregulated in WF. In BF, the c83433 (CHI) expression was consistent with that in WF, and c82232 (5MaT1) was highly expressed throughout the development period. The last group contained only one gene (c107855 (DFR), 4%), which was highly expressed throughout the developmental stages in WF but showed the opposite pattern in BF.
Comparison of expression profiles of anthocyanin biosynthesis transcription factors between BF and WF
Ten transcription factors, namely, MYB (c75274, c94823), bZIP (c93507), C2H2 (c71956, c84178), and HD-Zip (c101652, c13343, c39706, c67782, and c88135), were identified among the 261 DEGs. The heat map revealed that c101652, c75274, c71956, and c39706 expression showed poor repeatability between samples. The remaining six transcription factors showed a consistent trend in BF and WF, but expression of c13343, c88135, c94823, c67782, and c93507 in BF was higher than that in WF (Fig. 3).
Validation of the expression of genes related to anthocyanin biosynthesis in L. ruthenicum Murr.
The transcript abundances of 21 selected differentially expressed structural genes from the anthocyanin biosynthesis pathways and 6 transcription factors at the five coloration stages between BF and WF were analyzed by qRT-PCR to verify the reliability of transcription data and understand the relationship between gene expression and anthocyanin content in fruit. Compared with the three key periods of fruit coloration for the establishment of the transcriptome database, two additional stages were added before (S3-be, 25 days after anthesis) and after S3 (S3-af, 45 days after anthesis) to comprehensively reflect the expression trend, bringing the total to five developmental periods (Fig. S2). Figure 4 shows that in the ripening fruits of BF, the expression of the structural genes CHS (c105048), CHS (c89559), CHI (c101670), F3′5′H (c102345), DFR (c92376), ANS (c70865), UFGT (c87418), and UFGT (c103977) was significantly positively correlated with anthocyanin accumulation rate. The expression profiles of all these genes were highly similar, with maximum transcript levels noted at S3-be and/or S3 (Fig. 4). All of these genes were lowly expressed in WF, especially F3′5′H (c102345), UFGT (c103977), ANS (c70865), DFR (c92376), and CHS (c105048) with expression levels that were 2391, 119, 96, 85, and 25 times lower than those in BF at S3, respectively. The expression patterns of anthocyanin regulatory genes, including MYB, HD-Zip, and bZIP, were also investigated by qRT-PCR, and nothing noteworthy was found. The expression levels of all six transcription factors were low with minimal changes during development between BF and WF. Regulatory genes may not be responsible for color differences between BF and WF.
Discussion
In this study, the major biosynthetic pathways related to the fruit pigmentation of L. ruthenicum Murr. were investigated through transcriptomic analyses. The candidate genes that determined the biosynthesis and branch were isolated, and the regulatory mechanism of key genes was discussed.
The black pigmentation of ripe L. ruthenicum Murr. fruit is due to the high accumulation of specific anthocyanin (petunidin derivatives accounted for 95% of the total anthocyanins in fresh fruit that is derived from the delphinidin branch and mediated by F3′5′H activity) (Zheng et al. 2011). By contrast, extremely low anthocyanin was detected in the WFs of L. ruthenicum Murr. (Fig. 1a, b). The naturally white fruits provide an opportunity to unravel the complex molecular mechanism underlying certain color traits and to illustrate differences in anthocyanin accumulation between BF and WF.
The anthocyanin biosynthesis pathway is blocked and leads to the production of white L. ruthenicum Murr. berry. The expression levels of many genes in the pathway are inhibited. In particular, the expression levels of F3′5′H (c102345), UFGT (c103977), ANS (c70865), DFR (c92376), and CHS (c105048) were 2391, 119, 96, 85, and 25 times lower than those in BF at S3, respectively. This result strongly suggests that the low expression of these genes is responsible for the anthocyanin loss in WF. The anthocyanin biosynthesis pathway is usually divided into two parts: early and late sections (Deroles 2009). The early section leads to the formation of the dihydroflavonols, which comprises CHS, CHI, F3H, and CHS, the primary precursor for all classes of flavonoids. When CHS reactions are strongly constrained, the production of anthocyanin and nearly all other flavonoids is effectively eliminated (Clark et al., 2011). Hence, CHS mutation is the most common technique of inducing color loss (Clark and Verwoerd 2011; Saito et al. 2006; Spitzer et al. 2007). On the contrary, F3′H and F3′5′H catalyze the hydroxylation of the B-ring of flavonoids and are necessary to biosynthesize cyanidin (red to magenta) and delphinidin (violet to blue) based anthocyanins, respectively. As expected, the transcript level of three F3′H was low, and that of one F3′5′H was extremely high in BF. These findings are consistent with their funneling role for the metabolic flux toward the delphinidin-derived anthocyanin accumulation in L. ruthenicum Murr. instead of to the cyanidin-derived anthocyanin direction (Fig. 4, Zheng et al. 2011). All these genes are underexpressed in WF. The transcripts of F3′5′H-like sequence showed substantially higher levels in BF than in WF, particularly in S3 cases that are > 2391 times higher (Fig. 4). The delphinidin synthesis reactions are constrained to almost zero, but the metabolic flux does not induce the cyanidin accumulation in WF. DFR, ANS, and UFGT are key genes for anthocyanin formation and reduce the colorless leucoanthocyanidins to stabilize the colored compounds. In this study, the expression of DFR (c92376), ANS (c70865), and UFGT (c103977) in WF showed lower transcriptional abundance than in BF throughout the fruit development. DFRs have two variants: non-specific DFRs that convert all types of dihydroflavonols and specific DFRs that convert only dihydroquercetin (DHQ) and dihydromyricetin (DHM) (Miosic et al. 2014; Jin et al. 2016). Approximately 95% of anthocyanins in the fresh fruit of L. ruthenicum Murr. are petunidin (Zheng et al. 2011), indicating that the DFR in this plant is specific to only converting DHM and thus is similar to the DFR of gentians (Katsumoto et al. 2007), when it is co-expressed with F3′5′H, more efficient for metabolic flux toward delphinidin biosynthesis. Hence, the inhibited expression of these genes in WF may explain the lack of delphinidin. Furthermore, the suppression of the expression of regulatory genes may be the reason underlying the absence of structural genes. The mutation of genes encoding transcription factors that cause white flower formation has been reported in many species (Espley et al. 2009; Chiu et al. 2010; Jin et al. 2016). In the study, the expression patterns of anthocyanin regulatory genes (MYB, HD-Zip, and bZIP) were also investigated by qRT-PCR, and nothing noteworthy was found. The results revealed that the low expression of structural genes is not caused by the inhibition of regulatory genes and the regulatory genes are not responsible for the loss of anthocyanin in WF. Here, a hypothesis is proposed explaining the lack of color phenotype of L. ruthenicum Murr. fruits. The actual mechanism might be highly complicated. Thus, further elucidation and verification will be conducted through the quantitative analysis of compounds in the metabolic pathways of color substances and the transformation of key DEGs.
References
Altintas A, Kosar M, Kirimer N, Baser K, Demirci B (2006) Composition of the essential oils of Lycium barbarum, and L. ruthenicum fruits. Chem Nat Compd 42(1):24–25
Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor Vv MYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154:1470–1480. https://doi.org/10.1104/pp.110.164160
Clark ST, Verwoerd WS (2011) A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinformatics 12(1):343. http://www.biomedcentral.com/1471-2105/12/343
Deroles S. Anthocyanin biosynthesis in plant cell cultures: a potential source of natural colourants (2009) In: Kevin G, Kevin D, Chris W, editors. Anthocyanins: biosynthesis, functions and applications. pp 107–117. New York, USA: Springer Science+ Business Media, LLC; p. 107–117
Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183. https://doi.org/10.1105/tpc.108.059329
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Han YP, Vimolmangkang S, Soria-Guerra RE, Korban SS (2012) Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J Exp Bot 63:2437–2447
Jin X, Huang H, Wang L, Sun Y, Dai S (2016) Transcriptomics and metabolite analysis reveals the molecular mechanism of anthocyanin biosynthesis branch pathway in different Senecio cruentus cultivars. Front Plant Sci 7:1307. https://doi.org/10.3389/fpls.2016.01307
Katsumoto Y, Mizutani M, Fukui Y, Brugliera F, Holton T, Karan M et al (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48:1589–1600
Lou Q, Liu Y, Qi Y, Jiao S, Tian F, Jiang L, Wang Y (2014) Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J Exp Bot 65(12):3157–3164
Miosic S, Thill J, Milosevic M, Gosch C, Pober S, Molitor C, Ejaz S, Rompel A, Stich K, Halbwirth H (2014) Dihydroflavonol 4-reductase genes encode enzymes with contrasting substrate specificity and show divergent gene expression profiles in Fragaria species. PLoS One 9(11):e112707
Peng Q, Song J, Lv X, Wang Z, Huang L, Du Y (2012) Structural characterization of an arabinogalactan-protein from the fruits of Lycium ruthenicum. Journal of Agricultural & Food Chemistry 60(37):9424–9429
Potterat O (2010) Goji (Lycium barbarum and L. chinense): phytochemistry, pharmacology and safety in the perspective of traditional uses and recent popularity. Planta Med 76(1):7–19
Qi YY, Zhu CY, Chen JF, Liu GY, Yang ZHW, Chen WSH (2019) Comparative analysis of the quality and health-promoting compounds of two-shaped fruits of wild Lycium ruthenicum Murr from the Qinghai–Tibet Plateau. Acta Physiologiae Plantarum 41:101
Saito R, Fukuta N, Ohmiya A, Itoh Y, Ozeki Y, Kuchitsu K, Nakayama M (2006) Regulation of anthocyanin biosynthesis involved in the formation of marginal picotee petals in petunia. Plant Sci 170:828–834
Salvatierra A, Pimente P, Moya-León MA, Herrera R (2013) Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochem Lett 90:25–36
Spitzer B, Zvi MMB, Ovadis M, Marhevka E, Barkai O, Edelbaum O, Marton I, Masci T, Alon M, Morin S, Rogachev I, Aharoni A, Vainstein A (2007) Reverse genetics of floral scent: application of tobacco rattle virus-based gene silencing in Petunia. Plant Physiol 145:1241–1250
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Zeng S, Wu M, Zou C, Liu X, Shen X, Hayward A, Liu C, Wang Y (2014a) Comparative analysis of anthocyanin biosynthesis during fruit development in two Lycium species. Physiol Plant 150(4):505–516
Zeng S, Liu Y, Wu M, Liu X, Shen X, Liu C, Wang Y (2014b) Identification and validation of reference genes for quantitative real-time PCR normalization and its applications in Lycium. PLoS One 9(5):e97039. https://doi.org/10.1371/journal.pone.0097039
Zhang D, Xia T, Dang S, Fan G, Wang Z (2018) Investigation of Chinese wolfberry (Lycium spp.) germplasm by restriction site-associated DNA sequencing (RAD-seq). Biochem Genet 56:575–585
Zheng J, Ding C, Wang L, Li G, Shi J, Li H, Wang H, Suo Y (2011) Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Food Chem 126(3):859–865
Funding
This study was funded by the National Natural Science Foundation of China (Grant No: 31660221), Basic research project of Qinghai Province (Grant No. 2018-ZJ-713), and Construction Project for Innovation Platform of Qinghai Province (Grant No. 2017-ZJ-Y41).
Author information
Authors and Affiliations
Contributions
QYY and LXL initiated and designed the research. QYY, WHB, and GWY performed the experiments. QYY, SWJ, and DL analyzed the data. QYY and JLL revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Peter Nick
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, Y., Wei, H., Gu, W. et al. Transcriptome profiling provides insights into the fruit color development of wild Lycium ruthenicum Murr. from Qinghai–Tibet Plateau. Protoplasma 258, 33–43 (2021). https://doi.org/10.1007/s00709-020-01542-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-020-01542-9