Abstract
Grapevine trunk diseases (Eutypa dieback, esca and Botryosphaeria dieback) are caused by a complex of xylem-inhabiting fungi, which severely reduce yields in vineyards. Botryosphaeria dieback is associated with Botryosphaeriaceae. In order to develop effective strategies against Botryosphaeria dieback, we investigated the molecular basis of grapevine interactions with a virulent species, Neofusicoccum parvum, and a weak pathogen, Diplodia seriata. We investigated defenses induced by purified secreted fungal proteins within suspension cells of Vitis (Vitis rupestris and Vitis vinifera cv. Gewurztraminer) with putative different susceptibility to Botryosphaeria dieback. Our results show that Vitis cells are able to detect secreted proteins produced by Botryosphaeriaceae, resulting in a rapid alkalinization of the extracellular medium and the production of reactive oxygen species. Concerning early defense responses, N. parvum proteins induced a more intense response compared to D. seriata. Early and late defense responses, i.e., extracellular medium alkalinization, cell death, and expression of PR defense genes were stronger in V. rupestris compared to V. vinifera, except for stilbene production. Secreted Botryosphaeriaceae proteins triggered a high accumulation of δ-viniferin in V. vinifera suspension cells. Artificial inoculation assays on detached canes with N. parvum and D. seriata showed that the development of necrosis is reduced in V. rupestris compared to V. vinifera cv. Gewurztraminer. This may be related to a more efficient induction of defense responses in V. rupestris, although not sufficient to completely inhibit fungal colonization. Overall, our work shows a specific signature of defense responses depending on the grapevine genotype and the fungal species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trunk diseases are major grapevine diseases and a true challenge for viticulture. Eutypa dieback, esca, and Botryosphaeria dieback are harmful diseases caused by a complex of xylem-inhabiting fungi generating severe yield reduction in vineyards (for review, see Bertsch et al. 2013). The occurrence of these diseases has increased, leading to massive economic losses estimated to exceed one billion dollars per year (Hofstetter et al. 2012). Trunk diseases are generally associated with various symptoms, such as sectorial and/or central necrosis in woody tissues (Larignon 2012), brown stripes or canker (Rovesti and Montermini 1987; Larignon and Dubos 2001), and leaf discolorations and wilt of inflorescences and berries resulting in plant death (Larignon 2012). Grapevine trunk diseases are considered as major threat to winegrowers, since effective plant protection strategies are not available to date.
Mycelium of the causal fungi is detected in the trunk and other woody parts of the vines (cordons, spurs, canes), whereas it has never been isolated from symptomatic leaves or berries of infected plants (Larignon and Dubos 1997). It has been hypothesized that the fungi secrete toxins, which are translocated to the aerial organs via the xylem sap (Mugnai et al. 1999). While the symptoms and epidemiology of trunk diseases have been extensively studied in the past (for review, see Bertsch et al. 2013), the molecular and cellular mechanisms driving the interactions between host plant and fungal pathogens remain obscure. In this study, we focused on Botryosphaeria dieback, which is known to be associated with a wide range of Botryosphaeriaceae species (for review, see Úrbez-Torres 2011). Botryosphaeria dieback causes wood discolorations, gray sectorial necrosis, and orange/brown area just beneath the bark, dead spurs, and canes and foliar discolorations that vary from red to white cultivars, leading to premature plant death (for review, see Larignon 2012). Both in vitro and in vivo pathogenicity assays showed that Neofusicoccum parvum was one of the most virulent species whereas Diplodia seriata was one of the least virulent (Niekerk et al., 2004; Martos et al. 2008; Úrbez-Torres et al. 2008; Luque et al. 2009; Ramírez-Suero et al. 2014; Bénard-Gellon et al. 2015; Bellée et al. 2017).
Botryosphaeria dieback-associated fungi synthesize several virulence factors such as different metabolites inducing plant cell death (toxins), as well as hydrolytic enzymes responsible for wood degradation. Among the phytotoxic secondary metabolites produced by the main fungi associated with Botryosphaeria dieback, mellein, cis, trans-4-hydroxymellein, 4,7-dihydroxymellein, and naphthalenone-related compounds have been described (for review, see Andolfi et al. 2011). Moreover, Bellée et al. (2017) showed that an isolate of N. parvum (PER20) had characteristic metabolite profiles with the presence of salicylic acid derivatives and terremutin. Toxins also belong to the family of exopolysaccharides: a hydrophilic high-molecular-weight compound secreted by five Botryosphaeriaceae showed phytotoxic properties (Martos et al. 2008). Proteins were also identified as toxins in fungal invasion during trunk diseases. Bénard-Gellon et al. (2015) investigated the activity of secreted proteins produced by N. parvum and D. seriata on Vitis vinifera cv. Chardonnay calli. Compared to D. seriata, N. parvum Bourgogne S-116 produced extracellular proteins in higher quantity and with a higher biological activity, by inducing cell death in Vitis calli.
It is important to know if grapevine is able to rapidly recognize pathogens associated with trunk diseases and to identify the associated responses. The first line of plant defense is the recognition of conserved microbe molecules known as microbe-associated molecular pattern (MAMPs) through pattern recognition receptors (PRRs); for review, see Newman et al. 2013). This recognition leads to MAMP-triggered immunity (MTI) associated with the intracellular signal transduction and activation of early defense responses (Newman et al. 2013). MTI is characterized by ion flux modifications resulting in influxes of H+ and Ca2+ and effluxes of K+ and Cl− through the plasma membrane (Garcia-Brugger et al. 2006). H+ fluxes will trigger an extracellular alkalinization, which was proven toxic for the pathogen and associated with the activation of basal defense responses (for review, see Ebel and Scheel 1997). ROS production, also called oxidative burst, is another hallmark of MTI; it is toxic for the pathogen and involved in MAP kinase and defense gene activation, in cell wall reinforcement, and in hypersensitive reaction (for review, see Garcia-Brugger et al. 2006). The early signaling events will in turn lead to the production of secondary signals and phytohormones involved in the setup of local defense responses such as phytoalexin accumulation and PR protein synthesis, both having antimicrobial activity especially against fungi (for review, see Kissen and Bones 2012; Van Loon et al. 2006a). In addition to MAMP recognition, the plant immune system can also be activated by endogenous molecules released due to cellular damage or pathogen attack; these molecules are referred as damage-associated molecular patterns (DAMPs). In contrast to MAMPs which are derived from microorganisms, DAMPs are host cell derived and both initiate and perpetuate innate immune responses, helping the protection of damaged tissue (Choi and Klessig 2016). The perception of DAMPs (altered self) allows plant cells to recognize a greater diversity of pathogens (Boutrot and Zipfel 2017).
In this study, to know if grapevine cells are able to perceive directly or indirectly secreted proteins of Botryosphaeriaceae, we used the experimental model grapevine suspension cells that have been already used for defense response studies. This model was demonstrated as a reproducible model system for the study of early grapevine responses to infection with Phaeomoniella chlamydospora, a fungus responsible for esca. Lima et al. (2012) showed a biphasic oxidative burst, an increase in stilbene production and the induction of genes encoding PR proteins and stilbene synthase in Vitis vinifera cv. Vinhão cells after addition of Phaeomoniella chlamydospora biomass extract. Grapevine suspension cells were also used to study the responses to Botrytis cinerea, Eutypa lata, and Trichoderma atroviride, a biocontrol agent for grapevine trunk disease fungi (Dadakova et al. 2015; Mutawila et al. 2016).
We used cell cultures of Vitis vinifera cv. Gewurztraminer and Vitis rupestris, with putative different susceptibility to Botryosphaeriaceae based on wood assays. For these two genotypes, we studied defense reactions induced by secreted proteins produced by different isolates of N. parvum and D. seriata, which have differential virulence. We analyzed H+ fluxes via extracellular alkalinization, a rapid method to study the perception of a pathogen in suspension cells (Felix et al. 1993): oxidative burst, cell death, and phytoalexin production as well as defense gene expression.
Material and methods
Plant material
Suspension cell cultures of V. vinifera cv. Gewurztraminer were established from anthers and maintained in adapted liquid MS medium with half strength major salts (Maillot et al. 2006) and 0.5 mg L−1 nicotinic acid, 0.5 mg L−1 pyridoxine HCl, 0.1 mg L−1 thiamine HCl, 100 mg L−1 myo-inositol, 2 mg L−1 glycine, 0.1 mg L−1 6-benzylaminopurine (BAP), and 30 g L−1 sucrose, at pH 5.8. Suspension cell cultures of V. rupestris were established from leaves (Qiao et al. 2010) and kindly provided by Pr. Peter Nick (KIT, Karlsruhe, Germany). V. rupestris cells were maintained in liquid MS medium containing 4.3 g L−1 Murashige and Skoog basal salts (Duchefa™, The Netherlands), 30 g L−1 sucrose, 200 mg L−1 KH2PO4, 100 mg L−1 myo-inositol, 1 mg L−1 thiamine, and 0.2 mg L−1 2.4-dicholorophenoxy-acetic acid (2,4-D), at pH 5.8. Cells were subcultured every 7 days by transferring 20 mL of stationary cells into 80 mL of fresh medium in 250 mL Erlenmeyer flasks. Cells were propagated in the dark under shaking (120 rpm), at 25 °C for V. vinifera cv. Gewurztraminer cells and 26 °C for V. rupestris cells. For the different experiments, 50 mL of 6-day-old cells was used. Calli of V. vinifera cv. Gewurztraminer and V. rupestris were obtained from the same tissues as the suspension cells were subcultured every 2 weeks on the above described solid media in the dark at 25 ± 0.5 °C, with a 70 ± 10% relative humidity. For each tested condition, 15 calli with a diameter of 10 ± 2 mm were used. Inoculation experiments were conducted on detached canes, which consisted of 0.8 × 10 cm sections between two nodes of 1-year-old dormant lignified canes collected from Vitis vinifera cv. Gewurztraminer and Vitis rupestris. Both accessions were obtained from a collection in INRA Colmar, France.
Fungi
N. parvum isolate Bourgogne S-116 (Bourgogne, France), N. parvum isolate Bt-67 (Estremadura, Portugal), and D. seriata isolate 98.1 (Pyrénées Orientales, France) were isolated from vineyards. According to Ramírez-Suero et al. (2014), all fungi were grown on Petri dishes containing Potato Dextrose Agar (PDA) solid medium at 26 °C in the dark. After 10 days, the solid cultures were transferred in liquid medium by introducing three mycelia plugs (2.5 × 2.5 cm) from each fungus in 250 mL malt medium. Liquid cultures were grown at 220 rpm, at 28 °C in the dark. After 21 days, culture media were retrieved and sterilized by successive filtrations at 0.8, 0.45, and 0.2 μm.
Detached-cane inoculations
Inoculation experiments were conducted on detached canes, which consisted of 0.8 × 10 cm sections of 1-year-old shoots harvested from a collection from INRA Colmar, France (December 2015 and 2016, February 2017). Data were obtained from three independent experimental series of seven internodes for each genotype and fungi combination. A power drill no. 5 (Bosch PSB 500R) was used to wound the woody stems (5 mm in diameter, 3 mm in depth) at the center point between nodes. Cuttings were inoculated by filling the wound with a 6-mm-diameter plug collected from a 14-day-old fungal culture on PDA and then sealing this inoculation site with Para-film (American National Can). After inoculation, detached internodes were incubated in saturating humidity at 28 °C in darkness. After 21 days of incubation for D. seriata 98.1 and 7 days in the case of N. parvum Bt-67, the stems were debarked and necrotic areas were quantified from digital images using the ImageJ software (Schneider et al. 2012). We used a multifactorial ANOVA and a comparison of means using the Tukey test to compare the percentages of necrosed internode area between V. vinifera cv. Gewurztraminer and V. rupestris following inoculation with Neofusicoccum parvum Bt-67 (7 days post-inoculation) and Diplodia seriata 98.1 (14 days post-inoculation).
Extracellular protein preparation
Total extracellular proteins were precipitated from 250 mL filtered culture medium during 2 h at 4 °C with 60% (w/v) ammonium sulfate, according to Bénard-Gellon et al. (2015). After centrifugation (30 min at 10,000 rpm), the protein pellets were resuspended in deionized water and dialyzed in 3.5 kDa cutoff tubbing against deionized water for 20 h at 4 °C. Collected proteins were freeze-dried and resuspended in ultra-pure water. The protein concentration was measured with a BioSpec-nano Micro-volume Spectrophotometer (Shimadzu™, Japan) at 280 nm.
Measurement of alkalinization and ROS production in cell cultures
In order to determine the optimal protein concentration to apply, we first carried out a dose-response assay on both grapevine cell suspension cultures with various protein concentrations (10 to 200 μg mL−1) secreted by the highly virulent N. parvum Bourgogne S-116 strain (Ramírez-Suero et al. 2014; Bénard-Gellon et al. 2015). We determined that the protein concentration of 50 μg mL−1 is adequate to obtain a significant pH response in both cell suspensions (Online Resource 1).
Extracellular medium alkalinization was assessed by measuring the extracellular medium pH of 6-day-old suspension cultures after addition of extracellular proteins (50 μg mL−1). Before treatment, cells were equilibrated on an orbital shaker for 30 min. The pH value of the culture medium was recorded every 10 s for 30 min with a Handylab 12 pH meter (SI Analytics™, Germany), combined with a liquid electrolyte glass electrode (BlueLine Electrodes, SI Analytics™, Germany). In order to measure the pH increase, the pH of the extracellular medium at the beginning of the experiment (T0) was subtracted from all successive measures.
General oxidative stress response was assessed with the CM-H2DCFDA substrate (ThermoFisher™, USA). The day before the experiments, cells were filtered and resuspended in their respective culture medium at concentrations of 0.2 g fresh weight mL−1 for V. vinifera cv. Gewurztraminer and 0.4 g fresh weight mL−1 for V. rupestris. Then, 485 μL of the cell suspension was distributed in a 24-well plate and incubated with CM-H2DCFDA (ThermoFisher™, USA at a 1 μM final concentration. Four technical replicates were realized for each measure. After orbital shaking (1 mm) for 1 hour, extracellular proteins were dispensed at a 50 μg mL−1 concentration and plates were placed in a TriStar2 LB 942 Multidetection Microplate Reader (Berthold Technologies™, GmbH & Co. KG, Germany). The excitation filter was set at 485 nm, and the emission filter was set at 530 nm. The fluorescence from each well was measured and recorded by MikroWin software (Berthold Technologies™, GmbH & Co. KG, Germany). Data points were acquired each minute for 3 h. Data are mean and SD of four technical and two biological replicates.
Determination of cell viability
Determination of cell viability was assessed according to Lima et al. (2012). After treatment of a 6-day-old cell culture with extracellular fungal proteins for 24 and 48 h, 150 μL of cells was mixed with 50 μL of 0.4% (w/v) trypan blue (Sigma™, USA) and incubated in the dark for 15 min. Cells were then counted under a light microscope (×400; Alphaphot-2 YS2-H, Nikon®, Japan). Percentage of dead cells (stained blue) was calculated using the formula [(blue cells/total cell number) × 100]. Cell death was also assessed on V. vinifera cv. Gewurztraminer and V. rupestris calli. Fungal proteins were added to the callus medium at a concentration of 50 μg mL−1. Two-week-old Vitis vinifera cv. Gewurztraminer and V. rupestris calli were subcultured on the medium containing fungal proteins and incubated at 25 °C in the dark. The toxicity of secreted proteins was evaluated by visual observation of necrosis on calli at three time points (0, 3, and 6 days).
Measurement of stilbene production
Stilbene production was assessed by liquid chromatography–mass spectrometry. After treatment with extracellular proteins, cells were vacuum filtered at 3, 6, 24, and 48 h, frozen in liquid nitrogen, and stored at − 80 °C. Cells were ground in a mortar with liquid nitrogen, and then, methanol (5 mL mg−1 of fresh cells) and 5-methyl salicylate (1 μg) as an internal standard were added. Cells were agitated for 3 h at 4 °C with a Tube Rotator (Thermo Scientific™, USA). After centrifugation (2 × 20 min, 14,000 rpm), supernatant was collected and dried in a vacuum concentrator (Concentrator 5301, Eppendorf™, Germany), resuspended in a 40 μl methanol/water (90:10) solution, filtered at 0.2 μm, and kept at − 20 °C until LC-MS analysis. Trans-resveratrol, trans-piceid, 5-methylsalicylic acid, and trans-pterostilbene were purchased from Sigma-Aldrich™ (Saint-Quentin-Fallavier, France). Trans-δ-viniferin was purchased from Polyphenols Biotech™ (Villenave d’Ornon, France). Trans-ε-viniferin was kindly provided by Prof. Waffo-Téguo (University of Bordeaux). Cis-isomers of these five stilbenes were obtained by photoisomerization at 350 nm using a Rayonet photochemical reactor (Southern New England Ultraviolet Co.™). All solutions were prepared in 18 MΩ deionized water. LC-MS grade water, methanol, and formic acid were purchased from Fisher Scientific™ (Illkirch, France). The analytical system used was high-performance liquid chromatography Agilent 1100 series coupled with Agilent 6510 accurate-mass Quadrupole-Time of Flight (Q-TOF) mass spectrometer with ESI interface in negative ionization mode (Agilent Technologies™, California, USA). Data acquisition system software was Agilent MassHunter version B.02.00. An Agilent Zorbax C18 column (3.1 × 30 mm, 3.5 μm; Agilent Technologies™) was used at 35 °C, and the injected volume was 3 μL. The elution gradient was carried out with binary solvent system consisting of 0.1% formic acid in H2O (solvent A) and 0.1% formic acid in MeOH (solvent B) at a constant flow rate of 0.35 mL min−1. The gradient elution program was as follows: 0–4.0 min, 23–40% B, held for 2.0 min; 6.0–7.0 min, 40–70% B, held for 3.0 min; then up to 100% B; and kept at 100% of B during 4 min, followed by 4 min of stabilization at 23% B. The mass spectrometer operated by detection in the single ion monitoring (SIM) mode at the following settings: drying gas 13.0 L min−1 at 325 °C; nebulizer pressure 35 psi; capillary voltage − 3500 V; and fragmentor 150 V. Negative mass calibration was performed with standard mix G1969-85000 (Agilent Technologies™). Data processing was performed with Agilent MassHunter Qualitative and Quantitative software version B.07.00. Absolute contents of stilbenes were estimated from external calibration curves prepared with pure standards and internal standard (5-methylsalicylic acid) for extraction recovery.
Transcriptomic analysis
Defense gene expression was assessed by quantitative real-time (qRT) PCR analysis. After treatment with extracellular proteins, cells were vacuum filtrated at 3, 6, 24, and 48 h and stored at − 80 °C. RNA extraction, DNase treatment, and cDNA synthesis were achieved according to Bénard-Gellon et al. (2015). Cells were ground in liquid nitrogen, and total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen™, Germany) according to the manufacturer’s instructions and quantified using a BioSpec-nano Micro-Volume Spectrophotometer (Shimadzu™, Japan). Residual genomic DNA was removed by DNase1 digestion on extraction column with the RNase-free DNase set (Qiagen™, Germany) at 25 °C for 15 min; 1 μg of DNase-treated RNA was reverse transcribed with the iScript TM Reverse transcription supermix following the manufacturer’s instructions (Bio-Rad™, France). For real-time PCR, reactions were carried out on the CFX96 system (Bio-rad™, France). qPCR reactions were carried out in triplicates in a reaction buffer containing 2× iQ SYBR® Green Supermix, 0.2 mM of forward and reverse primers, and 10 ng of reverse transcribed RNA in a final volume of 25 μl. Thermal cycling conditions were as follows: 30 s at 95 °C followed by 40 cycles of 15 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C. The calibration curve for each gene was obtained by performing real-time PCR with serial dilutions of the purified PCR product (from 102 to 108 cDNA copy number). The specificity of the individual PCR amplification was checked using a heat dissociation curve from 55 to 95 °C following the final cycle of the PCR and by sequencing the final PCR products. The results obtained for each gene of interest were normalized to the expression of two reference genes, VvEF1α, an elongation factor 1α gene, and VvGAPDH, glyceraldehyde 3-phosphate dehydrogenase gene (Reid et al. 2006). Fold induction compared to appropriate controls (see legend of figures) was calculated as described by Hellemans et al. (2007). Mean values and standard deviations were obtained from three technical and three biological replicates. Primers used for quantitative real-time PCR are listed in Table 1.
Statistical analysis
Data were analyzed by using a multifactorial ANOVA and a multiple comparison of the means using the Tukey test (p ≤ 0.05) performed with R 3.3.2 software (R Development Core Team 2016). We used a multifactorial ANOVA to examine the effects of extracellular proteins from three fungal isolates (Neofusicoccum parvum Bourgogne S-116, Neofusicoccum parvum Bt-67, and Diplodia seriata 98.1) on ROS production, cell viability, defense gene expression, and stilbene production for cell lines of two plant genotypes (Vitis vinifera cv. Gewurztraminer and Vitis rupestris) at different time points according to the figure legends. Principal component analysis (PCA) was performed with the Mass Profiler Professional software (Agilent Technologies™, California, USA) where values are normalized by log2 transformation.
Results
Secreted Botryosphaeriaceae proteins induced rapid extracellular medium alkalinization in Vitis cells
Extracellular alkalinization is an early event following the perception of a pathogen molecular signal in suspension-cultured cells (Felix et al. 1993). Adding secreted proteins (50 μg mL−1 concentration) from the three Botryosphaeriaceae strains to grapevine cell suspensions induced a rapid increase in extracellular medium pH (Fig. 1). These results show that V. vinifera cv. Gewurztraminer and V. rupestris cells are able to respond to secreted proteins of fungi associated with Botryosphaeria dieback. Alkalinization intensity and velocity depends on both the fungus and the grapevine genotype, with N. parvum Bourgogne S-116 inducing the higher ΔpH on both Vitis cells, followed by N. parvum Bt-67 and D. seriata 98.1. For V. vinifera cv. Gewurztraminer cells (Fig. 1a), ΔpH increased around 7 min after addition of secreted proteins for N. parvum Bourgogne S-116, reaching a maximum of 0.5 unit above the initial pH (value of 4.5–5) within 30 min. For N. parvum Bt-67, the increase in ΔpH was recorded 9 min after treatment to reach a maximum of 0.4 after 30 min. Adding D. seriata 98.1 extracellular proteins led to a later and lower ΔpH, starting after 11 min and only reaching a maximum of 0.14 at 30 min. In V. rupestris cells (Fig. 1b), the initial pH was around 4.5–5, and proteins from the various fungi induced a faster and higher level of alkalinization. Compared to V. vinifera cv. Gewurztraminer, the pH increase started much earlier, from 2 to 3 min after treatment, to reach at 30 min a maximum ΔpH of 1 for N. parvum Bourgogne S-116, 0.93 for N. parvum Bt-67, and 0.7 for D. seriata 98.1. We further examined if the effect of the secreted fungal proteins on pH is due to their biological activity or tridimensional structure by adding heat denaturated proteins (50 μg mL−1) of the three fungi to both V. vinifera and V. rupestris suspension cells. No change in extracellular alkalinization was recorded with heat-denaturated proteins (data not shown).
a, b Extracellular medium alkalinization in V. vinifera cv. Gewurztraminer suspension cells (a) and V. rupestris (b) in response to secreted proteins at a 50 μg mL−1 final concentration of N. parvum Bourgogne S-116 (black lines), N. parvum Bt-67 (black dotted lines), and D. seriata 98.1 (gray dotted lines). Control cells (black dotted lines) were treated with water. Representative time lines from three independent series are shown
Secreted Botryosphaeriaceae proteins induced ROS production in Vitis cells
Production of ROS in Vitis cells was evaluated by the fluorescence measure of CM-H2DCFDA. The production of ROS was different according to both fungus and grapevine genotype (Fig. 2). N. parvum Bt-67 induced the higher ROS production in both Vitis cells, followed by N. parvum Bourgogne S-116 and D. seriata 98.1. In V. vinifera cv. Gewurztraminer cells (Fig. 2a), ROS production started about 11 min after addition of secreted proteins for N. parvum Bt-67, reaching a maximum at 83 min and decreasing thereafter. For N. parvum Bourgogne S-116 and D. seriata 98.1, ROS production was measured 21 min after treatment to reach a maximum at 120 and 145 min respectively. In V. rupestris cells (Fig. 2b), proteins from different fungi induced a faster but lower ROS production. ROS production started from 10 min after treatment and reached a maximum more rapidly compared to V. vinifera cv. Gewurztraminer, at 30 min for N. parvum Bourgogne S-116, 45 min for N. parvum Bt-67, and D. seriata 98.1. Maximum of relative fluorescence (258000) was measured with N. parvum Bt-67 (compared to approximately 1000 000 in V. vinifera cv. Gewurztraminer).
a, b Production of ROS measured by the fluorescence of CM-H2DCFDA in V. vinifera cv. Gewurztraminer suspension cells (a) and V. rupestris (b) in response to secreted proteins of N. parvum Bourgogne S-116 (gray dotted lines), N. parvum Bt-67 (black lines), and D. seriata 98.1 (light gray lines). Proteins were added to a 50 μg mL−1 final concentration. For each data points, values of water control were subtracted to values of protein treated cells. Each data point represents means of four technical and two biological replicates. Error bars represent the standard deviation of the mean. The asterisks indicate significant difference between treatments with the different fungal proteins at p ≤ 0.05 (Tukey Contrasts)
Secreted Botryosphaeriaceae proteins induced cell death only in V. rupestris cells
To determine if Botryosphaeriaceae extracellular proteins are able to induce cell death in Vitis cells, we followed cell viability with trypan blue staining at 24 and 48 h after treatment (Fig. 3). In V. vinifera cv. Gewurztraminer, no significant difference (p ≤ 0.236) in cell death was observed between protein and water control treatments (about 11% of cell death) (Fig. 3a). By contrast, in V. rupestris cells, all protein treatments induced cell death with significant (p ≤ 0.001) but various intensities, depending on the fungus. Extracellular proteins of N. parvum Bourgogne S-116 induced the highest cell mortality, 34% at 24 h and 40% at 48 h. N. parvum Bt-67 secreted proteins induced 18 and 23% of cell death at 24 and 48 h respectively, whereas D. seriata 98.1 caused a less intense cell death (15% at 24 h and 17% at 48 h) (Fig. 3b). To complete these results, we evaluated the toxicity of these secreted proteins on V. vinifera cv. Gewurztraminer (Fig. 3c) and V. rupestris (Fig. 3d) calli through necrosis observations. Proteins of each fungus were added to the callus culture medium at 50 μg mL−1, and their impact was observed after 3 and 6 days of contact. Partial necrosis was only observed after 3 days in V. rupestris with proteins from both N. parvum isolates. After 6 days, all V. rupestris calli in contact with secreted proteins showed partial necrosis (Fig. 3d). In contrast, secreted Botryosphaeriaceae proteins did not induce any necrosis in V. vinifera cv. Gewurztraminer.
a, d Induction of cell death in Vitis suspension cells by Botryosphaeriaceae proteins. Cell death assays were performed with cells of V. vinifera cv. Gewurztraminer (a) and V. rupestris (b) treated with secreted proteins (50 μg mL−1) of N. parvum Bourgogne S-116 (black bars), N. parvum Bt-67 (gray bars), and D. seriata 98.1 (light gray bars) compared to water control (white bars). Quantitative measurement of dead cells was performed by trypan blue staining at 24 and 48 h after treatment. Each data point represents means of three technical and two biological replicates. Error bars represent the standard deviation of the mean. Means with the same letter are not significantly different at p ≤ 0.05 (Tukey Contrasts). c, d V. vinifera cv. Gewurztraminer (c) and V. rupestris (d) calli subcultured for 6 days on MC+ medium supplemented with water (water control) or secreted fungal proteins. Pictures were taken after 0, 3, and 6 days of contact (dpi day post-inoculation)
Defense genes are differentially expressed in V. vinifera and V. rupestris cells in response to secreted Botryosphaeriaceae proteins
We further studied the expression of defense genes in V. vinifera cv. Gewurztraminer and V. rupestris suspension cells after treatment with secreted proteins from N. parvum and D. seriata (Fig. 4). These genes encode stilbene synthase (VvSTS1), a key enzyme involved in phytoalexin synthesis; superoxide dismutase (VvSOD), an enzyme involved in ROS scavenging and cellular detoxification; and several pathogenesis-related proteins (VvPR1, VvPR6, VvPR10.1, and VvChit4c) (Table 1). Our results show that three genes out of six in V. vinifera cv. Gewurztraminer (VvPR6, VvSTS1, and VvChit4c) and five genes in V. rupestris (VvPR1, VvPR6, VvPR10.1 VvSTS1, and VvChit4c) are induced by extracellular protein treatment. Overall, induction of defense genes is much stronger in V. rupestris cells, especially for VvPR6, VvChit4c, and VvPR10.1 (see the scale of graphs in Fig. 4).
a–f Defense gene expression (a VvPR1, b VvPR6, c VvPR10.1, d VvSTS1, e VvSOD, and f VvChit4C) in suspension cells of V. vinifera cv. Gewurztraminer (gray bars) and V. rupestris (black bars) treated with secreted proteins from N. parvum Bourgogne S-116, N. parvum Bt-67, and D. seriata 98.1. Gene expression was studied by RT-qPCR. The induction ratio represents the relative expression in cells treated with extracellular proteins (50 μg mL−1) versus control cells treated with water. Each data point represents the mean of three technical and three biological replicates. Error bars represent the standard deviation of the mean. Means with the same letter are not significantly different at p ≤ 0.05 (Tukey Contrasts)
VvPR1, encoding pathogenesis-related protein 1 is induced in response to a variety of pathogens and represents a useful molecular marker for the SAR response. In V. vinifera cv. Gewurztraminer, no significant difference in VvPR1 expression (p ≤ 0.436) was observed after addition of extracellular proteins from the different fungi (Fig. 4a). In V. rupestris cells, VvPR1 expression was slightly induced by N. parvum Bourgogne S-116 proteins at 3 and 6 h and highly induced by N. parvum Bt-67 proteins after 6 h (Fig. 4a).
VvPR6 encoding for a protease inhibitor is induced both in V. vinifera cv. Gewurztraminer and V. rupestris in response to Botryosphaeriaceae proteins (Fig. 4b). In V. vinifera cv. Gewurztraminer, induction of VvPR6 expression is significant at 24 h and maximal at 48 h (induction ratio of ∼ 30) for the two N. parvum isolates, while VvPR6 peaked at 24 h post-treatment with D. seriata 98.1 and decreased at 48 h. The induction of VvPR6 expression is significantly higher in V. rupestris cells, but only with the addition of N. parvum Bourgogne S-116 proteins: VvPR6 induction began as soon as 6 h and increased until 48 h post-treatment to reach a maximal induction ratio of ∼ 700.
VvPR10.1 was previously demonstrated to be upregulated in the interaction between V. vinifera Ugni blanc and Pseudomonas syringae (Robert et al. 2001). Expression of VvPR10.1 was only upregulated in V. rupestris cells, induction beginning at 6 h post-treatment for both N. parvum and D. seriata proteins. Upregulation of VvPR10.1 by D. seriata was lower compared to N. parvum (Fig. 4c).
VvChit4c encodes a class IV chitinase that could be involved in the fungal cell wall degradation. VvChit4c is the most upregulated gene in both V. vinifera cv. Gewurztraminer and V. rupestris cells, although upregulation was significantly higher in V. rupestris (Fig. 4f). Furthermore, the difference between the protein treatments is significant regardless of the genotypes (p ≤ 0.0001 for V. vinifera cv. Gewurztraminer and p ≤ 0.0001 for V. rupestris). The highest expression of VvChit4c was observed early, at 3 and 6 h post-treatment. In V. vinifera cv. Gewurztraminer, expression of VvChit4c was maximal 3 h after treatment with proteins of both N. parvum and D. seriata and decreased thereafter. In V. rupestris, expression of VvChit4c peaked 6 h post-treatment with all fungal proteins and decreased at later time points.
VvSTS1 encodes a stilbene synthase, a key enzyme in the production of phytoalexins. Compared to control cells, induction of VvSTS1 expression is much higher in V. rupestris cells compared to V. vinifera cv. Gewurztraminer. The intensity of VvSTS1 expression is also significantly different in both grapevine genotypes according to the different fungal protein treatment (Fig. 4d). Upregulation of VvSTS1 occurred as soon as 3 h post-treatment and was maximal with D. seriata proteins in V. vinifera cv. Gewurztraminer cells. In V. rupestris cells, proteins from N. parvum Bourgogne S-116 induced the highest expression of VvSTS1 with a peak at 6 h post-treatment.
VvSOD, which encodes a superoxide dismutase, is involved in ROS scavenging. VvSOD expression is not significantly affected by treatment with Botryosphaeriaceae proteins, although it is weakly and lately induced by N. parvum Bt-67 proteins at 48 h (Fig. 4e).
Overall, our results show that expression of several defense genes (VvPR6, VvChit4c, and VvSTS1) are more strongly upregulated in V. rupestris compared to V. vinifera cv. Gewurztraminer. In addition, some defense genes such as VvPR1 and VvPR10.1 are only induced in V. rupestris cells. Concerning the different fungi, N. parvum and especially N. parvum Bourgogne S-116 proteins were generally the most efficient in triggering defense responses compared to D. seriata, except for VvPR6 and VvSTS1 in V. vinifera cv. Gewurztraminer.
Secreted Botryosphaeriaceae proteins induced a different stilbene signature in V. vinifera and V. rupestris
In grapevine, the major phytoalexins belong to the family of stilbenes. We studied the production of resveratrol, δ- and ε-viniferins, pterostilbene, and piceid in suspension cells treated with Botryosphaeriaceae proteins (Fig. 5).
a–e Stilbenoid (a resveratrol, b piceid, c δ-viniferin, d ε-viniferin, and e pterostilbene) content in V. vinifera cv. Gewurztraminer (gray bars) and V. rupestris (black bars) at 3, 6, 24, and 48 h after treatment with secreted proteins (50 μg mL−1) from N. parvum Bourgogne S-116, N. parvum Bt-67, and D. seriata 98.1. Results represent the mean of three technical replicates and two biological replicates. Error bars represent the standard deviation of the mean. Means with the same letter are not significantly different at p ≤ 0.05 (Tukey Contrasts)
In control V. vinifera cv. Gewurztraminer cells, we observed a very low concentration of δ-viniferin (0.67 μg g−1 fresh weight (FW) after 24 h, Fig. 5c), pterostilbene (0.037 μg g−1 FW after 24 h, Fig. 5e), and ε-viniferin (0.006 μg g−1 after 24 h, Fig. 6d), whereas the concentration of resveratrol and piceid were relatively high (3.7 to 6.4 μg g−1 FW for resveratrol and 4.4 to 11.4 μg g−1 FW for piceid, Fig. 5a, b). Addition of N. parvum and D. seriata 98.1 proteins led to a similar stilbene pattern: levels of resveratrol tend to decrease (Fig. 5a), whereas there was a high increase in δ-viniferin overtime, reaching ∼ 15 μg g−1 FW at 24–48 h post-treatment (Fig. 5c). The levels of the other stilbenes (piceid, pterostilbene, and ε-viniferin) were not significantly affected by the addition of secreted Botryosphaeriaceae proteins.
Principal component analysis (PCA) of V. rupestris (black line) and V. vinifera cv. Gewurztraminer (black dotted line) stilbene profiles after inoculation with secreted proteins (50 μg mL−1) from N. parvum Bourgogne S-116 (triangles), N. parvum Bt-67 (circles), D. seriata 98.1 (squares), and water control (diamonds) in the two major components. Distribution of stilbene production defined by the first two axes (component 1 and component 2) was obtained with PCA of stilbenoid content using all data. The two major principal components explaining ~ 69% of the expression variance are plotted
Control V. rupestris cells had a different stilbene profile compared to V. vinifera cv. Gewurztraminer. Levels of resveratrol were lower (0.016 to 0.1 μg g−1 FW) whereas levels of piceid were higher (13.4 to 23.2 μg g−1). Treatment of V. rupestris cells with proteins from N. parvum induced a decrease in piceid, whereas after addition of D. seriata proteins, levels of piceid remained stable (Fig. 5b). Addition of proteins from N. parvum Bt-67 and D. seriata 98.1 induced a significant increase in δ- and ε-viniferin production at 24 and 48 h post-treatment. However, levels of ε-viniferin (app. 40 ng g−1 FW) were very low compared to δ-viniferin (app. 5 μg g−1 FW). Accumulation of δ-viniferin is also lower in V. rupestris compared to V. vinifera cv. Gewurztraminer (15 μg g−1 FW). In contrast to V. vinifera cv. Gewurztraminer, N. parvum Bourgogne S-116 proteins did not induce significant changes in δ-viniferin in V. rupestris. The levels of resveratrol and pterostilbene remained low and were not significantly affected by the addition of fungal proteins.
The difference in production of δ-viniferin, ε-viniferin, piceid, and resveratrol was found significantly different between V. vinifera cv. Gewurztraminer and V. rupestris cells regardless of the treatment with fungal proteins (p ≤ 0.001). Principal component analysis (PCA) used to visualize stilbene profile between the two genotypes further indicated that the first two principal components (component 1 and component 2) explained 69% of the total data variability (Fig. 6), confirming that stilbene profiles differed significantly according to the genotype.
In summary, our results show a different stilbene profile between V. vinifera cv. Gewurztraminer and V. rupestris. In comparison to control, the main difference is the accumulation of δ-viniferin in the two genotypes at a high level, which could indicate an important role of this stilbene in the defense response of Vitis cells.
Levels of necrosis induced by artificial internode inoculation with N. parvum and D. seriata are lower in V. rupestris compared to V. vinifera cv. Gewurztraminer
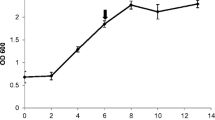
To know if the differential induction of defense responses in V. rupestris and V. vinifera cv. Gewurztraminer that we observed in suspension cells could be related to a different susceptibility to Botryosphaeria dieback associated fungi, we artificially inoculated 1-year-old canes from both genotypes with N. parvum and D. seriata. The area of necrosis was measured with ImageJ software 7 days post-inoculation for N. parvum and 14 days post-inoculation for D. seriata (Fig. 7). For both fungi, the area of necrosis was slightly smaller in V. rupestris compared to V. vinifera cv. Gewurztraminer (p ≤ 0.05). The area of necrosis was also higher after inoculation with N. parvum Bt-67 compared to D. seriata 98.1, showing that this fungus has a higher capacity to rapidly colonize grapevine wood.
Development of N. parvum (a) and D. seriata (b) in V. vinifera cv. Gewurztraminer and V. rupestris 1-year-old internodes after artificial inoculation. The percentage of necrosed area relative to total internode area was calculated for each internode using ImageJ software 7 days after inoculation for N. parvum and 14 days after inoculation for D. seriata. Mean ± SE was calculated from three independent experiments each comprising 7–8 internodes. The asterisks indicate significant difference between V. vinifera and V. rupestris at p ≤ 0.05 (Tukey contrasts)
Discussion
The molecular dialog between grapevine cells and fungi associated with trunk diseases remains poorly understood, although it is essential to determine if plant cells can perceive the invader and set up appropriate defense responses. Most of the studies on defense responses induced by fungi associated with trunk diseases have been conducted on vineyard plants, showing differential expression of foliar symptoms (Bertsch et al. 2013). Host-pathogen field trials are complex since several pathogens are involved. Moreover, field studies are affected by a number of environmental factors such as other biotic/abiotic stresses and climatic conditions. In this respect, controlled inoculations either using greenhouse cuttings or plantlets have been used as standard assays. It is important to notice that artificial wood inoculation with trunk disease fungi induces a wound response that can mask any effect of pathogen perception on gene induction at early times after infection (Pierron et al. 2016). In the present study, the simplified model we developed allows to better understand the molecular dialog between grapevine and trunk disease fungi.
Non-self-perception in plant innate immunity initially relies on the recognition of microbe-associated molecular patterns (MAMPs) (Boller and Felix 2009; Dodds and Rathjen 2010). Perception of pathogen attack can also be realized indirectly through the perception of DAMPs (altered self), which are endogenous molecules released by the host stressed cells (Boutrot and Zipfel 2017). MAMP and DAMP-triggered immunity results in the activation of a wide range of defense responses and confers a basal level of resistance to pathogens. We first investigated whether Vitis cells were able to perceive secreted proteins produced by Botryosphaeriaceae and to subsequently activate early defense responses. Extracellular alkalinization is an essential component of ion fluxes involved in plant defense and has been used as an efficient method to monitor chemosensory perception in cultured plant cells (Felix et al. 1993).
Several studies have shown that grapevine cells are able to react to several MAMPs such as chitin, rhamnolipids, and peptides such as flagellin and harpin (Delaunois et al. 2014). Following perception, early defenses such as ion fluxes and ROS production were activated but reports on such responses in the case of the interaction between grapevine and fungi associated with trunk diseases, especially Botryosphaeriaceae, are scarce. Our results show that both V. vinifera cv. Gewurztraminer and V. rupestris cells are able to recognize secreted proteins of fungi associated with Botryosphaeria dieback and respond very rapidly with modification of ion fluxes and ROS production. Since no difference in ΔpH was observed when heat-denaturated proteins of both N. parvum and D. seriata were applied on Vitis cells, we propose that induction of defense responses by these Botryosphaeriaceae proteins is not due to the recognition of a peptide motif. Defense induction could be triggered by fungal protein biological activity (through the release of DAMPs) or result from the recognition of MAMP tridimensional structure as previously shown for elicitins from Pythium oligandrum (Takenaka et al. 2011). In another previous study, Bénard-Gellon et al. (2015) also showed that heat-denaturated proteins from N. parvum failed to induce necrosis on Vitis vinifera calli, in contrast to native proteins. In addition, several DAMPs identified in the literature and recognized by plants belong to the protein family (for example systemin, subtilase, or ATP synthase (Boutrot and Zipfel 2017)).
It is important to point out that the extracellular medium alkalinization was more rapid and intense in V. rupestris cells compared to V. vinifera cv. Gewurztraminer. A higher and faster extracellular medium alkalinization was also measured in Vitis rupestris cells after challenge with harpin, a bacterial elicitor, compared to V. vinifera cv. Pinot Noir (Qiao et al. 2010). It has also been reported that elicitation of cell cultures of Vitis vinifera cv. Vinhão, an esca tolerant grapevine, with a crude extract of P. chlamydospora induced a biphasic oxidative burst, which may be characteristic of an incompatible reaction (Lima et al. 2012). The monophasic oxidative burst observed in our cells may indicate a compatible interaction between Vitis cells and Botryosphaeriaceae fungi. Overall, our results show that early defenses are triggered more rapidly in V. rupestris compared to V. vinifera cv. Gewurztraminer.
Since proteins produced by Botryosphaeria species induce cell death in V. rupestris, they could also represent toxins promoting fungal invasion in plant organs. For instance, in Eutypa dieback, proteins of variable molecular weight (6 to 200 kDa) are secreted by Eutypa lata and have been assumed to induce physiological imbalance resulting in cellular damages (Octave et al. 2006a, b). Similarly, a set of proteins secreted by Phaeomoniella chlamydospora and Phaeoacremonium minimum triggered cell death and transient H+ flux modifications in grapevine 41B cells (Luini et al. 2010). The authors demonstrated that H+-ATPase is targeted by these polypeptides, resulting in its inhibition. Another study demonstrated that V. rupestris responded to the harpin elicitor with a massive hypersensitive response (HR) type of cell death occurring within 48 h, whereas no cell death was observed in V. vinifera cv. Pinot Noir (Chang et al. 2011). HR cell death thus seems to be a hallmark of the defense responses in North American Vitis (Chang et al. 2011). In our system, Botryosphaeriaceae proteins induce cell death only in V. rupestris cells and not in V. vinifera cv. Gewurztraminer, and a higher oxidative burst was measured in Gewurztraminer compared to V. rupestris, which may seem contradictory. Indeed, massive ROS production under biotic and abiotic stresses is generally thought to trigger plant cell death. However, relation between ROS production and cell death is very complex and still a matter of debate (De Pinto et al. 2012). It was also previously shown that timing of ROS production (which is much more rapid in V. rupestris), as well as interaction with antioxidant systems, is very important for cell death induction (De Pinto et al. 2012). In our system, it remains to be further determined the role of ROS in cell death induction, which kind of cell death is triggered by our fungal proteins (necrosis or apoptotic HR cell death), and whether cell death favors fungal progression or contributes to plant resistance.
The rapid response of Vitis cells to secreted Botryosphaeriaceae proteins is in accordance with other studies using artificially inoculated whole plants as model. Recent RNA-Seq analysis revealed that inoculation of N. parvum triggered the expression of 20 candidate genes, which were rapidly induced in the leaves within minutes of inoculation and persisted throughout the disease latent phase, in the absence of infection development or change in wood anatomy (Czemmel et al. 2015). In another study, characterization of the transcriptional dynamics of grapevine genes in the woody stem and in the leaves during Neofusicoccum parvum colonization revealed that both stems and leaves undergo extensive transcriptomic reprogramming in response to infection of the stem, especially 24 h after inoculation (Massonnet et al. 2017). Analysis of the early perception of P. chlamydospora and P. minimum by measuring early expression of defense genes further suggested that grapevine perceives esca pathogens (Pierron et al. 2016). From all these studies, it appears that grapevine has the ability to sense very rapidly trunk disease invaders.
Our results further demonstrate that differences in early defense response activation in the two grapevine genotypes are further transduced into qualitative and quantitative differences in cell death, defense gene induction, and secondary metabolite signature. Concerning expression of defense markers, a higher number of defense genes are activated in V. rupestris compared to V. vinifera cv. Gewurztraminer and to a significant higher level. In V. rupestris, addition of secreted Botryosphaeriaceae proteins triggered the induction of PR1, PR6, PR10.1, Chit4C, and STS1, whereas in V. vinifera, only PR6, Chit4C, and STS1 are activated. The timing and level of induction also depend on the Botryosphaeriaceae strain, with N. parvum generally inducing defense genes to a higher level compared to D. seriata. Induction of defense markers is mostly rapid, beginning as soon as 3–6 h post-treatment. In another study, elicitation of grapevine cells with P. chlamydospora crude extract resulted in a rapid (3 h post-treatment) increase in the expression of PR6, PR10, β-1,3-glucanase, class III chitinase, lipoxygenase, phenyalanine ammonia lyase, and stilbene synthase (Lima et al. 2012). At the whole plant scale, the expression of PR protein genes was upregulated in the leaves of grapevines affected by Eutypa dieback and grapevine leaf stripe disease (Valtaud et al. 2009b; Camps et al. 2010; Letousey et al. 2010; Magnin-Robert et al. 2011; Spagnolo et al. 2012). In Botryosphaeria dieback affected vines, a proteome comparison of the brown striped wood showed the abundance of PR proteins (PR2, PR5, and PR17) and members of the antioxidant system (GST5, cysPEROX) in the brown striped wood of three V. vinifera cultivars (Spagnolo et al. 2014). An interesting study focused on defense responses induced by absorbed culture filtrates from P. minimum and P. chlamydospora on cuttings from grapevine cultivars showing differential susceptibility to esca disease (Lambert et al. 2013). Genes encoding PR proteins (GLU, PIN, CHIT4C, and PGIP) are highly stimulated in the less susceptible cultivars after fungal filtrate treatment. Accumulation of PR proteins could represent an important line of defense towards trunk disease fungi in V. rupestris, since a fungi-toxic activity has been described for many PR proteins (Van Loon et al. 2006a). Moreover, PR proteins generally show a greater accumulation in inoculated resistant plants compared with susceptible ones and a strong constitutive accumulation of PR proteins in plants with a high level of natural disease resistance is observed (for review, see Edreva 2005).
We also analyzed stilbene production in both grapevine suspension cells after treatment with secreted fungal proteins. Stilbenes are a family of secondary metabolites derived from the phenylpropanoid pathway and produced by a number of plants. Stilbenes have been implicated in both inducible and constitutive plant defenses to bioagressors, in a number of studies (Chong et al. 2009). In our system, synthesis of stilbenes and especially of the potent antifungal compound δ-viniferin was activated in both genotypes by all fungal strains. However, V. rupestris cells were characterized by a constitutive higher level in piceid, and lower levels in δ-viniferin were measured after treatment with fungal proteins compared to V. vinifera. In addition, accumulation of ε-viniferin, although to low levels compared to δ-viniferin, was only measured in V. rupestris following treatment with secreted fungal proteins. These results contrast with those from Chang et al. (2011), showing that V. rupestris produced massive δ-viniferin compared to V. vinifera cv. Pinot Noir after treatment with the bacterial elicitor harpin. However, it is possible that perception of different MAMPs by different receptors results in a different stilbene profile. We can also notice that resveratrol and piceid contents, respectively, in V. vinifera cv. Gewurztraminer and V. rupestris control cells, tend to decrease after addition of fungal proteins, suggesting that these two stilbenes could be further metabolized in other active forms such as viniferins.
Another point is that stilbene synthase expression is higher in V. rupestris compared to V. vinifera cv. Gewurztraminer after treatment with fungal proteins and this does not result in higher stilbene levels in this genotype. One possibility is that stilbenes are secreted into the extracellular medium, where they could be degraded by extracellular fungal enzymes or by reacting with ROS. Another possibility is that activation of stilbene synthase, responsible for resveratrol synthesis, is not the limiting factor resulting in stilbene accumulation, since resveratrol is further metabolized in other stilbenes.
Our results show that δ-viniferin accumulation could be an important component of grapevine response to Botryosphaeria dieback fungi. Other studies suggest the implication of stilbenes in the grapevine responses to trunk disease fungi. For example, hydroxystilbenes such as resveratrol and δ-viniferin, as well as more complex stilbenoids accumulate in brown red wood of grapevines showing esca symptoms (Amalfitano et al. 2011). Resveratrol and δ-viniferin also accumulate in grapevine plants after artificial inoculation with P. minimum and P. chlamydospora (Martin et al. 2009). Two phenylpropanoid biosynthesis genes (PAL) and stilbene synthase (STS) were highly expressed in asymptomatic leaves before the appearance of the apoplectic esca form (Letousey et al. 2010). Concerning the inhibiting activity of stilbenes on trunk disease fungi, a direct antifungal effect by inhibiting the in vitro growth of E. lata, S. hirsutum, and F. mediterranea was shown after application of resveratrol (Mazzullo et al. 2000). In a recent work, Lambert et al. (2012) evaluated the impact of 24 grapevine phenolic compounds on major wood decay fungi by in vitro agar plate assay. This study revealed that stilbenoids were the most active on fungal growth inhibition, especially on the Botryosphaeriaceae N. parvum and D. seriata. Among the tested compounds, piceatannol, ε-viniferin, and pterostilbene were the most actives, whereas δ-viniferin was not tested. Resveratrol oligomer content was also significantly higher in wood after inoculation of foliar cuttings with N. parvum, in contrast to D. seriata that did not induce significant changes in stilbenoids (Lambert et al. 2012). In our suspension cells, N. parvum and D. seriata proteins triggered a similar stilbene profile (Fig. 5).
In summary, our work shows that perception of proteins secreted by Botryosphaeriaceae by both grapevine genotypes is likely to be related to a biological activity or tridimensional structure and results in different signature of early and late defense responses depending on both grapevine genotype and fungal strain. V. rupestris is characterized by higher medium alkalinization, cell death, and more intense induction of PR genes, whereas V. vinifera cv. Gewurztraminer shows a higher production of antifungal δ-viniferin. A major question when using simplified model systems, such as cell cultures, is to define to what extent the results can be transposed to a whole plant, even if the molecular responses we studied probably occur. Indeed, based on artificial internode inoculation, we show that V. rupestris is somewhat more tolerant than V. vinifera cv. Gewurztraminer based on the area of necrosis in the wood. It is possible that lower fungal development in V. rupestris is related to cell death and higher PR induction. Comparison of defense responses in cultured cells versus inoculated wood could be performed in the future. However, it is important to point out that even if grapevine is able to sense rapidly trunk disease invaders, induction of defense responses is seemingly not strong enough to contain pathogen growth, since both N. parvum and D. seriata develop in the wood of both grapevine genotypes. N. parvum has been shown as much more aggressive compared to D. seriata (for review, see Úrbez-Torres 2011), although we show here that its secreted proteins are also more efficient in inducing grapevine defense responses. Generally, D. seriata, which was shown as a fungus with low aggressiveness, also induced the lower ion fluxes, ROS production, and expression of defense genes. But it is also important to consider that Botryosphaeriaceae fungi likely possess different enzymatic equipment that could explain wood and phytoalexin degradation, as well as toxin synthesis, resulting in the bypass of plant defense and leading to fungal progression.
As a conclusion, our work contributes to a better understanding of different Vitis genotype responses to Botryosphaeriaceae fungi and future studies will focus on aggressiveness factors (enzymatic degradation of wood and phytoalexins), as well as on the identification and characterization of secreted proteins and their mode of action.
Abbreviations
- qRT-PCR:
-
Reverse transcription quantitative real-time PCR
- SOD :
-
Superoxide dismutase
- STS1 :
-
Stilbene synthase 1
- PR1 :
-
Pathogenesis-related protein 1
- PR6 :
-
Pathogenesis-related protein 6
- PR10.1 :
-
Pathogenesis-related protein 10.1
- GLU :
-
Glucanase
- Chit4c :
-
Chitinase 4c
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- EF1α :
-
Elongation factor 1α
- PGIP :
-
Polygalacturonase-inhibiting protein
- PIN :
-
Serine protease inhibitor
- MAMPs:
-
Microbe-associated molecular pattern
- PRRs :
-
Pattern recognition receptors
- MTI:
-
MAMP-triggered immunity
- ROS:
-
Reactive oxygen species
- PR protein:
-
Pathogenesis-related protein
- HR :
-
Hypersensitive response
- LC-MS:
-
Liquid chromatography–mass spectrometry
- FW:
-
Fresh weight
- SAR:
-
Systemic acquired resistance
References
Amalfitano C, Agrelli D, Arrigo A et al (2011) Stilbene polyphenols in the brown red wood of Vitis vinifera cv. Sangiovese affected by “esca proper”. Phytopathol Mediterr 50:224–235. https://doi.org/10.14601/Phytopathol_Mediterr-9720
Andolfi A, Mugnai L, Luque J et al (2011) Phytotoxins produced by fungi associated with grapevine trunk diseases. Toxins 3:1569–1605. https://doi.org/10.3390/toxins3121569
Bellée A, Comont G, Nivault A, et al (2017) Life traits of four Botryosphaeriaceae species and molecular responses of different grapevine cultivars or hybrids. Plant Pathol 66:763–776. doi: https://doi.org/10.1111/ppa.12623
Bénard-Gellon M, Farine S, Goddard ML et al (2015) Toxicity of extracellular proteins from Diplodia seriata and Neofusicoccum parvum involved in grapevine Botryosphaeria dieback. Protoplasma 252:679–687. https://doi.org/10.1007/s00709-014-0716-y
Bertsch C, Ramírez-Suero M, Magnin-Robert M et al (2013) Grapevine trunk diseases: complex and still poorly understood. Plant Pathol 62:243–265. https://doi.org/10.1111/j.1365-3059.2012.02674.x
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. https://doi.org/10.1146/annurev.arplant.57.032905.105346
Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286. https://doi.org/10.1146/annurev-phyto-080614-120106
Camps C, Kappel C, Lecomte P et al (2010) A transcriptomic study of grapevine (Vitis vinifera cv. Cabernet-Sauvignon) interaction with the vascular ascomycete fungus Eutypa lata. J Exp Bot 61:1719–1737. https://doi.org/10.1093/jxb/erq040
Chang X, Heene E, Qiao F, Nick P (2011) The phytoalexin resveratrol regulates the initiation of hypersensitive cell death in Vitis cell. PLoS One 6:e26405. https://doi.org/10.1371/journal.pone.0026405
Choi HW, Klessig DF (2016) DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol 16:232. https://doi.org/10.1186/s12870-016-0921-2
Chong J, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177:143–155. https://doi.org/10.1016/j.plantsci.2009.05.012
Czemmel S, Galarneau ER, Travadon R et al (2015) Genes expressed in grapevine leaves reveal latent wood infection by the fungal pathogen Neofusicoccum parvum. PLoS One 10:e0121828. https://doi.org/10.1371/journal.pone.0121828
Dadakova K, Havelkova M, Kurkova B et al (2015) Proteome and transcript analysis of Vitis vinifera cell cultures subjected to Botrytis cinerea infection. J Proteome 119:143–153. https://doi.org/10.1016/j.jprot.2015.02.001
De Pinto MC, Locato V, De Gara L (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35:234–244. https://doi.org/10.1111/j.1365-3040.2011.02387.x
Delaunois B, Farace G, Jeandet P et al (2014) Elicitors as alternative strategy to pesticides in grapevine? Current knowledge on their mode of action from controlled conditions to vineyard. Environ Sci Pollut Res Int 21:4837–4846. https://doi.org/10.1007/s11356-013-1841-4
Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548. https://doi.org/10.1038/nrg2812
Ebel J, Scheel D (1997) Signals in host-parasite interactions. In: Carroll PDGC, Tudzynski PDP (eds) Plant relationships. Springer, Berlin Heidelberg, pp 85–105
Edreva A (2005) Pathogenesis-related proteins: research progress in the last 15 years. Gen Appl Plant Physiol 31:105–124
Felix G, Regenass M, Boller T (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J 4:307–316. https://doi.org/10.1046/j.1365-313X.1993.04020307.x
Garcia-Brugger A, Lamotte O, Vandelle E et al (2006) Early signaling events induced by elicitors of plant defenses. Mol Plant-Microbe Interact 19:711–724. https://doi.org/10.1094/MPMI-19-0711
Hellemans J, Mortier G, Paepe AD et al (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. https://doi.org/10.1186/gb-2007-8-2-r19
Hofstetter V, Buyck B, Croll D et al (2012) What if esca disease of grapevine were not a fungal disease? Fungal Divers 54:51–67. https://doi.org/10.1007/s13225-012-0171-z
Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17:73–90. https://doi.org/10.1016/j.tplants.2011.11.002
Lambert C, Bisson J, Waffo-Téguo P et al (2012) Phenolics and their antifungal role in grapevine wood decay: focus on the Botryosphaeriaceae Family. J Agric Food Chem 60:11859–11868. https://doi.org/10.1021/jf303290g
Lambert C, Khiook ILK, Lucas S et al (2013) A faster and a stronger defense response: one of the key elements in grapevine explaining its lower level of susceptibility to esca? Phytopathology 103:1028–1034. https://doi.org/10.1094/PHYTO-11-12-0305-R
Larignon P (2012) Maladies cryptogamiques du bois de la vigne: Symptomatologie et agents pathogènes. http://www.vignevin.com/menu-haut/actualites/article.html?tx_ttnews%5Btt_news%5D=368&tx_ttnews%5BbackPid%5D=918&cHash=2c0eccd030. Accessed 19 Jan 2017
Larignon P, Dubos B (1997) Fungi associated with esca disease in grapevine. Eur J Plant Pathol 103:147–157. https://doi.org/10.1023/A:1008638409410
Larignon P, Dubos B (2001) Le Black Dead Arm: Maladie nouvelle à ne pas confondre avec l’esca. Phytoma 538:26–29
Letousey P, Baillieul F, Perrot G et al (2010) Early events prior to visual symptoms in the apoplectic form of grapevine esca disease. Phytopathology 100:424–431. https://doi.org/10.1094/PHYTO-100-5-0424
Lima MRM, Ferreres F, Dias ACP (2012) Response of Vitis vinifera cell cultures to Phaeomoniella chlamydospora: changes in phenolic production, oxidative state and expression of defence-related genes. Eur J Plant Pathol 132:133–146. https://doi.org/10.1007/s10658-011-9857-4
Luini E, Fleurat-Lessard P, Rousseau L et al (2010) Inhibitory effects of polypeptides secreted by the grapevine pathogens Phaeomoniella chlamydospora and Phaeoacremonium aleophilum on plant cell activities. Physiol Mol Plant Pathol 74:403–411. https://doi.org/10.1016/j.pmpp.2010.06.007
Luque J, Martos S, Aroca A et al (2009) Symptoms and fungi associated with declining mature grapevine plants in northeast Spain. J Plant Pathol 91:381–390
Magnin-Robert M, Letousey P, Spagnolo A et al (2011) Leaf stripe form of esca induces alteration of photosynthesis and defence reactions in presymptomatic leaves. Funct Plant Biol 38:856–866. https://doi.org/10.1071/FP11083
Maillot P, Kieffer F, Walter B (2006) Somatic embryogenesis from stem nodal sections of grapevine. Vitis 45:185–189
Martin N, Vesentini D, Rego C et al (2009) Phaeomoniella chlamydospora infection induces changes in phenolic compounds content in Vitis vinifera. Phytopathol Mediterr 48:101–116. https://doi.org/10.14601/Phytopathol_Mediterr-2879
Martos S, Andolfi A, Luque J et al (2008) Production of phytotoxic metabolites by five species of Botryosphaeriaceae causing decline on grapevines, with special interest in the species Neofusicoccum luteum and N. parvum. Eur J Plant Pathol 121:451–461. https://doi.org/10.1007/s10658-007-9263-0
Massonnet M, Figueroa-Balderas R, Galarneau ERA, et al (2017) Neofusicoccum parvum colonization of the grapevine woody stem triggers asynchronous host responses at the site of infection and in the leaves. Front Plant Sci 8:1117. doi: https://doi.org/10.3389/fpls.2017.01117
Mazzullo A, Cesari A, Osti F, Di Marco S (2000) Bioassays on the activity of resveratrol, pterostilbene and phosphorous acid towards fungi associated with esca of grapevine. Phytopathol Mediterr 39:357–365
Mugnai L, Graniti A, Surico G (1999) Esca (black measles) and Brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis 83:404–418. https://doi.org/10.1094/PDIS.1999.83.5.404
Mutawila C, Stander C, Halleen F et al (2016) Response of Vitis vinifera cell cultures to Eutypa lata and Trichoderma atroviride culture filtrates: expression of defence-related genes and phenotypes. Protoplasma 254:863–879. https://doi.org/10.1007/s00709-016-0997-4
Newman M-A, Sundelin T, Nielsen JT, Erbs G (2013) MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4:139
Octave S, Amborabé B-E, Fleurat-Lessard P et al (2006a) Modifications of plant cell activities by polypeptides secreted by Eutypa lata, a vineyard fungal pathogen. Physiol Plant 128:103–115. https://doi.org/10.1111/j.1399-3054.2006.00715.x
Octave S, Roblin G, Vachaud M, Fleurat-Lessard P (2006b) Polypeptide metabolites secreted by the fungal pathogen Eutypa lata participate in Vitis vinifera cell structure damage observed in Eutypa dieback. Funct Plant Biol 33:297–307. https://doi.org/10.1071/FP05230
Pierron RJG, Pouzoulet J, Couderc C, et al (2016) Variations in early response of grapevine wood depending onwound and inoculation combinations with Phaeoacremonium aleophilum and Phaeomoniella chlamydospora. Front Plant Sci 7:268. doi: https://doi.org/10.3389/fpls.2016.00268
Qiao F, Chang X-L, Nick P (2010) The cytoskeleton enhances gene expression in the response to the Harpin elicitor in grapevine. J Exp Bot 61:4021–4031. https://doi.org/10.1093/jxb/erq221
R Development Core Team (2016) R: a language and environment for statistical computing. Austria, Vienna
Ramírez-Suero M, Bénard-Gellon M, Chong J et al (2014) Extracellular compounds produced by fungi associated with Botryosphaeria dieback induce differential defence gene expression patterns and necrosis in Vitis vinifera cv. Chardonnay cells. Protoplasma 251:1417–1426. https://doi.org/10.1007/s00709-014-0643-y
Reid KE, Olsson N, Schlosser J et al (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27. https://doi.org/10.1186/1471-2229-6-27
Robert N, Ferran J, Breda C et al (2001) Molecular characterization of the incompatible interaction of Vitis Vinifera leaves with Pseudomonas syringae pv. pisi: expression of genes coding for stilbene synthase and class 10 PR protein. Eur J Plant Pathol 107:249–261. https://doi.org/10.1023/A:1011241001383
Rovesti L, Montermini A (1987) Un deperimento della vite causato da Sphaeropsis malorum diffuso in provincia di Reggio Emilia. Inf Fitopatol 37:59–61
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671
Spagnolo A, Magnin-Robert M, Alayi TD et al (2012) Physiological changes in green stems of Vitis vinifera L. cv. Chardonnay in response to esca proper and apoplexy revealed by proteomic and transcriptomic analyses. J Proteome Res 11:461–475. https://doi.org/10.1021/pr200892g
Spagnolo A, Magnin-Robert M, Alayi TD et al (2014) Differential responses of three grapevine cultivars to Botryosphaeria dieback. Phytopathology 104:1021–1035. https://doi.org/10.1094/PHYTO-01-14-0007-R
Takenaka S, Yamaguchi K, Masunaka A et al (2011) Implications of oligomeric forms of POD-1 and POD-2 proteins isolated from cell walls of the biocontrol agent Pythium oligandrum in relation to their ability to induce defense reactions in tomato. J Plant Physiol 168:1972–1979. https://doi.org/10.1016/j.jplph.2011.05.011
Úrbez-Torres JR (2011) The status of Botryosphaeriaceae species infecting grapevines. Phytopathol Mediterr 50:5–45. https://doi.org/10.14601/Phytopathol_Mediterr-9316
Úrbez-Torres JR, Leavitt GM, Guerrero JC et al (2008) Identification and pathogenicity of Lasiodiplodia theobromae and Diplodia seriata, the causal agents of bot canker disease of grapevines in Mexico. Plant Dis 92:519–529. https://doi.org/10.1094/PDIS-92-4-0519
Valtaud C, Foyer CH, Fleurat-Lessard P, Bourbouloux A (2009) Systemic effects on leaf glutathione metabolism and defence protein expression caused by esca infection in grapevines. Funct Plant Biol 36:260–279. https://doi.org/10.1071/FP08293
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162. https://doi.org/10.1146/annurev.phyto.44.070505.143425
van Niekerk JM, Crous PW, Groenewald JZ(E) et al (2004) DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia 96:781–798
Acknowledgements
E. Stempien PhD fellowship was financed by the Alsace Region (France) and CIVA (Conseil Interprofessionnel des Vins d’Alsace). V. rupestris cells were a kind gift of Pr. Peter Nick (Karlsruher Institut fur Technologie, Karlsruhe, Germany) and? trans-ε-viniferin which was kindly provided by Prof. Waffo-Téguo (University of Bordeaux). We are grateful to D. Merdinoglu and V. Dumas for taking samples of woods of V. vinifera cv Gewurztraminer and V. rupestris in INRA Colmar. This work has been supported by Interreg Rhin Supérieur projects “Bacchus” and “Vitifutur.”
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Hanns H. Kassemeyer
Electronic supplementary material
ESM 1
(DOCX 89 kb)
Rights and permissions
About this article
Cite this article
Stempien, E., Goddard, ML., Leva, Y. et al. Secreted proteins produced by fungi associated with Botryosphaeria dieback trigger distinct defense responses in Vitis vinifera and Vitis rupestris cells. Protoplasma 255, 613–628 (2018). https://doi.org/10.1007/s00709-017-1175-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-017-1175-z