Abstract
Since its discovery, atomic force microscopy (AFM) is widely used to study biological objects and materials, including cells, proteins, and nucleic acids. AFM measurements are carried out in the air as well as in liquid with a very high resolution, even more complex bioprocesses can be monitored in situ under physiological conditions. Successful imaging of DNA molecules on the flat supporting surface typically requires appropriate treatment of mica. The original surface charge of mica is the same as of DNA, i.e. negative. Accordingly, immobilization using bivalent cations (Mg2+, Ni2+, and Co2+), deposition of ethanolamine, and mica surface silanization with alkoxysiloxane derivatives were reported to achieve an optimal concentration and surface arrangement of DNA molecules. Vapours of alkoxysiloxane derivatives led to uniform negatively charged mica surface and it was found that higher ionic radius causes a weaker bond. A better quality and sharper images of DNA molecules were achieved by adjusting the correct real amplitude of the cantilever. This amplitude should correspond with the expected size of the target objects—DNA molecules in the x–y plane of the image. The length of the observed DNA molecules was 1000 bp and the planar width of DNA was 7.8 nm (in reality 3 nm). The AFM spectroscopic mode was particularly useful.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atomic force microscopy (AFM) belongs to scanning probe microscopic (SPM) techniques; this experimental method was introduced in 1986 by G. Binnig, C. F. Quate, and Ch. Gerber [1–4]. The key idea leading to AFM was utilization of systematic variations accompanying measurements with the scanning tunnelling microscope (STM), which is monitoring the current between the conductive tip and a conductive sample. As measurement of non-conductive samples was required, a novel concept focused directly on the atomic forces and soon microscope based on the atomic forces—AFM was constructed [5–7]. AFM employs a flexible cantilever with sharp probe (tip) scanning the sample, a cantilever deflection results from force interactions between the probe and the imaged surface [8–10]. While scanning, the tip moves in contact or in a close proximity over the sample and it becomes either attracted or repulsed by surface atoms of the sample [5]. AFM employs laser beam focused on the cantilever from which it is reflected onto a 4-quadrant photodetector. The change of tip position caused by interaction with the sample is detected from the deflection of the cantilever, converted to a voltage error signal which is used in a feedback loop to control elongation of piezoceramic elements [8, 11–13] driving the probe. The resulting topographic map of sample is usually accompanied by additional information about surface properties.
AFM provides topographic images with nanometer resolution, sufficient for visualization of individual biological molecules as proteins and nucleic acids usually deposited on the atomically flat and clean surface of mica. Mica provides very flat surface that can be modified for immobilization of biomolecules. For the first time, DNA was imaged on mica by Bustamante and colleagues in 1994 by contact mode in air [14]. They also introduced the treatment of mica with magnesium ions for weak adsorption of DNA. Divalent ions of Mg2+ functioned as bridges between DNA and mica surface. Further progress addressed studies of DNA dynamics in solution. Imaging using contact mode in fluids was difficult (capillary forces and weak adsorption of DNA on mica). Major break came with introduction of the tapping mode that allowed reproducible imaging of DNA both in air and even in fluid; interactions of DNA with specific molecules were also followed [15]. Few years later, treatment of mica surface with 3-aminopropyltriethoxysilane (APTES) was introduced for stronger adsorption of DNA. This method is applied in different variations up to present and some modifications are presented here.
In this contribution, several immobilisation methods leading to strong DNA attachment on mica are compared in order to obtain more realistic data better corresponding to the expected dimensions of DNA. The optimization steps providing better resolution and contrast in the tapping mode of DNA based on magnitude-height dependences are discussed.
Results and discussion
AFM topography of DNA
For immobilization of DNA on negatively charged surfaces including mica, suitable bivalent cations usually play an important role. When mica becomes exposed to aqueous solution of bivalent ions, it undergoes ion-exchange process in which potassium ions naturally contained on its surface are replaced by other ions from the solution. The divalent ions can create interlinks between the negatively charged mica and the negative phosphate groups of DNA. In this way, DNA is confined by electrostatic forces to the surface of mica. Later, it was found that mica need not be preincubated with divalent ions, but for a strong binding, addition of divalent ions directly into the buffer with DNA is sufficient [16–19].
The ion radius of individual atoms was quite significant as illustrated in Fig. 1, which shows AFM topography of calf thymus double-stranded DNA (dsDNA) immobilized on the mica surface with the help of either Mg2+, Co2+, or Ni2+. The comparison of average cross-sections of DNA obtained from ten profile measurements, is available in Table 1.
From the obtained AFM topography, in case of magnesium, DNA was bound on mica through the positive charge of Mg2+ (Fig. 1a), but clearly the interaction was not strong enough to keep DNA on mica in sufficient quantity corresponding to the applied concentration. Immobilization of DNA was not completely successful, even though the experiment was carefully optimised, gradual exposure of mica to initially 1 mM MgCl2 was done, gentle washing and drying followed, and finally dsDNA (1 μg cm−3) dissolved in 1 mM MgCl2 was applied. The imaging results confirmed the expected effect of the ionic radius on the strength of the interactions of DNA [17]. The reason for the reduced efficiency of DNA binding to mica was caused by the rather large ionic radius of Mg2+ (72 pm). Furthermore, the open places on mica not covered by DNA were partially occupied by salt crystals (visible in the image as white spots), which was another disadvantage of immobilization of DNA with the help of magnesium ions. The measured cross-section of dsDNA was 17.5 nm.
For the alternative immobilization of DNA (concentration was always 1 μg cm−3), it was directly diluted in 1 mM solution of either NiCl2 (Fig. 1b) or CoCl2 (Fig. 1c) without previous surface modification, strong binding interactions were confirmed. The resulting AFM images of DNA were in terms of binding interactions with both bivalent ions substantially similar. DNA topological features were better established compared to the use of Mg2+ ions due to the lower ion radius of both Ni2+ (69 pm) and Co2+ (65 pm). Average sizes of DNA in these cases were 17.6 nm (Co2+) and 16.7 nm (Ni2+), data shown in Table 1.
Alternative method of modification of mica for subsequent immobilization of DNA employed ethanolamine; it has been used to modify the cantilever tip for force spectroscopy elsewhere [2]. Here, this method was adapted for use with mica for AFM topography. The obtained topographic images of DNA are shown in Fig. 2. The binding interaction was very strong; unfortunately, there was also evident artefact due to the cracked AFM tip, i.e. doubled, slightly out of focus of the image of DNA chain. Anyway, the measured average cross-section of DNA was 10.8 nm, especially with the help of appropriately chosen values of real amplitude of cantilever.
Finally, immobilization and imaging of DNA was also tested on the silanized mica (Fig. 3). From the chemical point of view, this is a process that allows covalent immobilization of biomolecules on the surface of the carrier. It provides activation of mica by a contact layer of silane with a suitable terminal amino group; modification was achieved using the evaporative deposition [20]. The most common reagent is 3-(2-aminoethylamino)propyltriethoxysilane (APTES), the resulting product is sometimes referred to as AP-mica (AP-aminopropyl). The obtained amino groups have pK A about 10.6 and even though accumulation of amino groups on the surface of AP-mica decreases with decreasing pH, the surface in solution at neutral pH remains positively charged. DNA again binds to the AP-mica surface through electrostatic interactions [16].
AFM topography (scan size 1 × 1 μm) of the dsDNA fragments from calf thymus. dsDNA was immobilized on mica surface with the method “vapour” APTES/APDMES in a desiccator at first without alkaline catalysis a only “vapour” APTES silanization, b APDMES. In both cases, there was undesirable self-polymerization. Then “vapour” silanization was also used c for APTES with the help of strong alkaline-triethylamine
If a monolayer of silane is desirable, then 3-(aminopropyl)dimethylethoxysilane (APDMES) is chosen; it cannot generate lateral cross-links and complex structures often resulting due to the self-polymerization of e.g. APTES (typically deposited as 4–5 layers of silane) in the presence of water. Utilization of APDMES is also appropriate with an organic catalyst. APDMES can form hydrogen bonds with hydroxyl groups on the surface of mica with both ends of its molecule: ethoxy and aminopropyl groups. During the silanization reaction, the quantity of silane associated with the mica surface through Si–O–Si bond gradually increases primarily by hydrogen bond of the ethoxy group with the hydroxyl group of surface. Aminopropyl groups of silane are less reactive, but basically block the surface and thereby limit the desired level of binding. Therefore, for a faster and more quantitative silanization, alkaline catalysis is used as it increases the number of amine groups protruding from the surface [21].
For most cases, the vapour-based deposition of both APTES (Fig. 3a) and APDMES (Fig. 3b) and also APTES combined with alkaline catalysis (Fig. 3c) provided surfaces unsuitable for immobilization of DNA; silanization resulted in poorly defined surfaces coated with oligomerized silane multilayers. The only successful exception was the combination of APDMES and alkaline catalysis with TEA where reasonable imaging of DNA was possible (Fig. 5). In the former cases (Fig. 3), despite making mica preparation in dry conditions of a desiccator, where it was protected from moisture known to support polymerization of silanes, the generated surfaces were not suitable for imaging of DNA. It was confirmed that even the shortest minimal manipulation with silanized mica during application of the DNA sample exposed the surface to humidity and this stimulated undesired polymerisation and prevented topography of the examined DNA. In the case of silanization with APDMES under the basic catalysis of triethylamine, it was confirmed that presence of a single ethoxy groups of in APDMES was significant. Only this surface was reasonably flat and strongly binding DNA.
Effect of real amplitude of cantilever for imaging
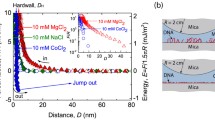
For the best imaging of biomolecules, the amplitude of oscillation of the cantilever plays a significant role for sharpness of borders and correct dimensions evaluation of the examined objects. Information on the absolute value of the amplitude can be obtained from the magnitude-height plot from the AFM spectroscopy (Fig. 4). From these curves, the values of real amplitudes of cantilever were determined as 38.3 and 7.9 nm. A higher amplitude of cantilever (Fig. 4a) provided unfocused image and a larger cross section of the DNA compared to reality. An amplitude value below 7.9 nm (Fig. 4b) has not yet been achieved with this type of cantilever—inferred according to the size of a particular peak in the resonant spectrum and signal noise. For precise imaging of small objects, the amplitude of the cantilever oscillations should be comparable to the expected dimensions. When the amplitude is significantly larger, the topographic images provide poor resolution and incorrect measurements. This is demonstrated for DNA (expected cross-section of 2–3 nm), where the large amplitude at 38.3 nm resulted in a blurred image (Fig. 5a) and it provided the size of DNA equal to 19.6 nm, i.e. far away from reality. On the other hand, much finer amplitude of 7.9 nm (Fig. 5b) resulted in a nicely resolved DNA shapes and cross-section size of 7.8 nm which is much more realistic.
Conclusions
From the time of the invention of AFM, DNA represents one of the most studied biological molecules. The important reasons for imaging of DNA include characterization of it surface properties, detection of possible damages, location and quantification of damaged sites either directly or with the help of antibodies and other DNA interacting proteins. In future, this effort can help with diagnosis of diseases and provide a better understanding of tumour growth. The results presented in this paper focused on the initial, often rather underestimated step in such research—immobilization of DNA on mica surface. Attachment of DNA on mica is not a straightforward process; at physiological pH, mica exhibits a negative charge similarly as negatively charged phosphate groups in the DNA chain. Thus, modification of the mica surface by introducing a positively charged contact interlayer should be initially performed and several such techniques were compared.
The positive surface charge can be obtained through the introduction of bivalent metal ions; the ionic radius of the ions was significant for strong interaction with DNA. The suitable ionic radius complements the charge based binding of DNA and thus provides higher forces for formation of DNA-surface links. Alternatively, coatings providing amino group were considered, including either simple deposition of ethanolamine or formation of silane layers with terminal amino groups, providing positive charge at suitable pH of the working buffer. This method, however, was successful only when 3-aminopropyldimethylethoxysilane was applied under strong alkaline catalysis; its molecule contains only a single alkoxy-group and thus it was suitable for lateral cross-linking of the silanes resulting in undefined layers not suitable for AFM imaging. In this case, the most realistic images were achieved after fine tuning of the real amplitude of cantilever providing cross-section of DNA 7.8 nm.
Experimental
The sample preparation methods described below are applicable for imaging DNA on mica surface. Mica is commonly used as a carrier for AFM molecular imaging resolution due to its atomically flat surface [16, 22].
Properties and modification of mica
For AFM measurements, Mica Grade V-1 Muscovite (2SPI Supplies, USA) was used. This substrate is highly popular in the field of AFM, mainly due to its perfect cleavage, flexibility, and chemical resistance. In terms of chemical structure, it is a compound of K2O·Al2O3·SiO2 and its surface can be considered as hydrated SiO2, therefore chemically well modifiable. The surface is highly hydrophilic and negatively charged at physiological pH (pK A ~ 3). Immobilization is carried out either by ionic interactions or covalent binding of biomolecules [23, 24].
To achieve a clean surface, fresh layer of mica was cleaved using the scotch tape. After cleavage, potassium cations are distributed on mica surface, leaving uncompensated negative charges. Therefore, freshly cleaved negatively charged mica is an excellent support for positively charged molecules. The adsorption of negatively charged molecules, such as DNA, requires special sample preparation procedures [16], as described below.
Bivalent cations
DNA from calf thymus, length about 1000 bp (Sigma-Aldrich, USA) was prepared for measurement in the solution of the chosen ion. DNA was always diluted to the final concentration of 1 μg cm−3. For deposition, three types of ions were used, MgCl2, CoCl2, and NiCl2 all at 1 mM concentration. This mixture was then placed on the freshly cleaved mica surface, incubated for 30 min and gently dried with the stream of air in case of CoCl2 and NiCl2. In case of Mg2+ cations, 5 mm3 of stock solution of 1 mM MgCl2 was pipetted on the freshly cleaned mica surface. Mica was then placed for 10 min into a refrigerator in a Petri dish. After the incubation period, mica was rinsed and dried with compressed air. DNA (1 μg cm−3 diluted in 1 mM MgCl2) was then pipetted on mica treated with Mg2+ cations and allowed to dry completely in air. The site of application was rinsed once again with distilled water [16].
Ethanolamine/HCl
Modification was carried out with ethanolamine (5 mg cm−3) dissolved in dimethylsulfoxide (DMSO). This mixture was placed in a beaker together with molecular sieve (aluminosilicate, Ø 1.6–2.5 mm) and freshly cleaved mica and heated at 100 °C overnight. The next day, mica was washed three times alternately in DMSO and ethanol and dried with compressed air [2, 25]. Afterwards, the stock solution of DNA was diluted to 0.5 μg cm−3 in 40 mM HEPES and 10 mM MgCl2 buffer. DNA solution was then placed on the mica surface, incubated for 30 min, and dried with compressed air [16].
Silanization
Freshly cleaved mica sheets were placed in Petri dish in desiccator and the open tube with 30 mm3 of either APTES (Fluka, Switzerland) or APDMES (Sigma-Aldrich, USA) was placed in the desiccator which was closed and vacuum was applied for 10 min. This evaporative silanization yielded AP-mica in 2 h [16].
For the AP-mica procedure with alkaline catalysis, another tube with 30 mm3 of triethylamine (TEA, Fluka, Switzerland) was inserted into desiccator [21]. The next step was immobilization of DNA with concentration 0.5 μg cm−3 diluted in 40 mM HEPES/10 mM MgCl2 buffer. DNA solution was then placed on the mica surface, incubated for 30 min and dried with compressed air [16].
AFM measurement
NTEGRA-Vita atomic force microscope (NT-MDT, Zelenograd, Russia) was used in the tapping mode and all measurements were performed in air environment. Special sharp AFM probes—ETALON, HA-NC (NT-MDT) were used; these provide tips made of silicon and with radius of curvature close to 10 nm. For each measurement, average value (from ten measurements) of cross-section of DNA was also calculated using the evaluation software Gwyddion 2.27. For improved imaging, for each new AFM probe the real amplitude of cantilever was determined using the AFM spectroscopy; the magnitude MAG (nA) versus height (nm) plots were generated and the real amplitude of cantilever was calculated using the formula: AMPreal/nm = (dX/dY) × SetPoint/nA, where SetPoint corresponds to the half of the free oscillation amplitude of cantilever [26].
References
Santos NC, Castanho MARB (2004) Biophys Chem 107:133
Riener CK, Stroh CM, Ebner A, Klampfl C, Gall AA, Romanin C, Lyubchenko YL, Hinterdorfer P, Gruber H (2003) Anal Chim Acta 479:59
Lyubchenko Y, Shlyakhtenko L, Harrington R, Oden P, Lindsay S (1993) Proc Natl Acad Sci USA 90:2137
Wickramasinghe HK (2000) Acta Mater 48:347
Jalili N, Laxminarayana K (2004) Mechatronics 14:907
Ikai A (1996) Surf Sci Rep 26:261
Yin Y, Zech M, Williams TL, Hoffman JE (2009) Phys C 469:535
Alessandrini A, Facci P (2005) Meas Sci Technol 16:R65
Fotiadis D, Scheuring S, Müller SA, Engel A, Müller DJ (2002) Micron 33:385
Jandt KD (2001) Surf Sci 491:303
Butt HJ, Cappella B, Kappl M (2005) Surf Sci Rep 59:1
Braga PC, Ricci D (2004) Atomic force microscopy: biomedical methods and applications. Humana Press, New Jersey
Alonso JL, Goldmann WH (2003) Life Sci 72:2553
Ando T, Uchihashi T, Fukuma T (2008) Prog Surf Sci 83:337
Bhushan B, Kawata S (2006) Applied scanning probe methods VI: characterization. In: Thomson NH (ed) Atomic force microscopy of DNA structure and interactions. Springer, Berlin, p 127
Lyubchenko YL, Shlyakhtenko LS, Ando T (2011) Methods 54:274
Bezanilla M, Manne S, Laney DE, Lyubchenko YL, Hansma HG (1995) Langmuir 11:655
Zheng J, Li Z, Wu A, Zhou H (2003) Biophys Chem 104:37
Sun XG, Cao EH, Zhang X, Liu D, Bai C (2002) Inorg Chem Commun 5:181
Sasou M, Sugiyama S, Yoshino T, Ohtani T (2003) Langmuir 19:9845
White LD, Tripp CP (2000) J Colloid Interface Sci 232:400
Costa LT, Pinto JR, Moraes MB, De Souza GGB, Sorenson MM, Bisch PM, Weissmuller G (2004) Biophys Chem 109:63
Campbell PA, Sinnamon LJ, Thompson CE, Walmsley DG (1998) Surf Sci 410:L768
Liu Z, Li Z, Zhou H, Wei G, Song Y, Wang L (2005) Micron 36:525
Ouerghi O, Touhami A, Othmane A, Ben Ouada H, Martelet C, Fretigny C, Jaffrezic-Renault N (2002) Sens Actuators B Chem 84:167
Thormann E, Pettersson T, Kettle J, Claesson PM (2010) Ultramicroscopy 110:313
Acknowledgments
This work was supported by Central European Institute of Technology (CZ.1.05/1.1.00/02.0068) from European Regional Development Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Horňáková, V., Přibyl, J. & Skládal, P. Study of DNA immobilization on mica surface by atomic force microscopy. Monatsh Chem 147, 865–871 (2016). https://doi.org/10.1007/s00706-016-1695-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-016-1695-9