Abstract
Cereal chlorotic mottle virus (CCMoV) is a cicadellid-transmitted plant rhabdovirus associated with chlorotic and necrotic streaks on several gramineous hosts and weeds. The virus was initially described in 1979 in Australia, but its genome has never been sequenced. In this study, the complete genome sequence of a Moroccan isolate of CCMoV was generated by high-throughput sequencing from infected oat leaves (Avena sativa). The genome is 13,800 nt long, containing seven open reading frames (ORFs) arranged in the canonical organization of rhabdoviruses: 3’-nucleocapsid (N), phosphoprotein (P), unknown protein (p3), unknown protein (p4), matrix (M), glycoprotein (G), viral polymerase (L)-5’. Pairwise analysis showed that maize fine streak virus (MFSV, genus Gammanucleorhabdovirus) was the closest relative. The amino acid identity values between homologous proteins from CCMoV and MFSV are as follows: 59.27% (N), 36.7% (P), 24% (P3), 62% (P4), 43.70% (M), 49.15% (G), 60.93% (L). Based on its phylogenetic relationship and analogous genome architecture, CCMoV should be assigned as member of the genus Gammanucleorhabdovirus. The low sequence similarity observed between CCMoV and MFSV suggests that CCMoV is a member of a distinct virus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Members of the family Rhabdoviridae have a negative-sense, single-stranded RNA genome of 10-16 kb in length and a typical bacilliform or bullet-shaped virion morphology, although filamentous virions have also been reported [1]. The natural host range for this viral family encompasses several kingdoms, including vertebrates, invertebrates, and plants. Rhabdoviruses from plants and arthropod vectors are grouped in the subfamily Betarhabdovirinae, which contains six genera: Alphanucleorhabdovirus, Betanucleorhabdovirus, Gammanucleorhabdovirus, Cytorhabdovirus, Dichorhavirus, and Varicosavirus [1].
Cereal chlorotic mottle virus (CCMoV) is an unclassified plant rhabdovirus that was initially described in 1979 in Australia (CCMoV-A) [2]. CCMoV was later reported in Morocco (CCMoV-M) in 1985 [3] and detected infecting barley in Spain in the 1990s (Lockhart, unpublished data). The host range of CCMoV includes several economically important gramineous hosts and common weeds, including Avena sativa, Digitaria ciliaris, Dinebra retroflexa, Echinochloa colona, Eleusine coracana, Eleusine indica, Hordeum vulgare, Triticum aestivium, Setaria verticillata, Agrostis semiverticillata, Phalaris sp., Triticum durum, Oryzopsis miliacea, and some cultivars of corn (Zea mays) [2, 3]. As for transmission, two cicadellid insect vectors have been identified: the Australian grass leafhopper (Nesoclutha pallida) for CCMoV-A and the orange leafhopper (Cicadulina bipunctata subsp. bipunctella) for CCMoV-M [2,3,4]. Purified virus particles of CCMoV have the typical rhabdovirus morphology, with bullet-shaped or bacilliform virions, and ultrastructural studies had shown their accumulation in the perinuclear space and cytoplasmic vesicles of infected plant cells [3, 4]. However, the taxonomic classification of CCMoV among the plant-infecting rhabdoviruses is still pending due to the lack of its genomic sequence.
In the present study, the complete genome sequence of the original Moroccan isolate of CCMoV (CCMoV-M) was determined. A freeze-dried sample of oat (Avena sativa) infected with CCMoV-M, stored at the University of Minnesota Virology Laboratory since 1985, was used as the virus source. Rhabdovirus-like particles were confirmed by electron microscopy (TEM) of crude sap preparations. The sample was fixed with 2.5% glutaraldehyde (GA) and negatively stained with 2% phosphotungstic acid (PTA) (Fig. 1A) as described previously [3]. Total RNA was extracted from 100 mg of freeze-dried tissue using an RNeasy Plant Mini Kit (QIAGEN, Germany), and residual DNA was removed by DNase I digestion. Ribosomal RNA (rRNA) was depleted using a Ribo-Zero Plant Kit (Illumina, USA). A complementary DNA (cDNA) library was generated using a TruSeq Stranded Total RNA Library Prep Kit and sequenced on a NextSeq 500 Illumina platform. Sequence reads were processed using Geneious Prime® 2022.0.1 [5]. The BBDuk plugin was used during the preprocessing step to remove adapter and low-quality reads [6]. Sequencing yielded 44,109,524 raw data reads (75-bp single-end reads) after quality trimming. The sequence reads were assembled de novo using the SPAdes assembler, resulting in 103,842 contigs. A BLAST search (NCBI) identified a viral contig of 13,371 nt sharing the highest level of identity 69% at the nucleotide level (coverage, 40%) with maize fine streak virus (MFSV). No other viral contigs were identified.
A Virus particle of cereal chlorotic mottle virus (CCMoV-M) from infected oat leaves. Scale bar 100 nm. B Alignment of gene junctions detected in CCMoV; all differences from the consensus sequence are highlighted in black. IUPAC ambiguity code W (A/U). Three motifs were separated based on homology to MFSV [9], those correspond to the 3’ end of mRNAs (column 1), intergenic sequences (column 2), and the 5’ ends of the mRNAs (column 3). C Diagram depicting the genome arrangement of cereal chlorotic mottle virus (CCMoV-M). Putative ORFs are represented by grey boxes. The asterisk (*) indicates the relative position of the gene junctions. The nucleotide sequences observed at each junction are detailed in panel B of this figure
To obtain the complete genome sequence of the rhabdovirus, the RNA termini were sequenced using a SMARTer® RACE 5’/3’ Kit (Takara, USA) according to manufacturer instructions, with some minor modifications to obtain the 3’-end sequence. The template RNA used for the 3’-RACE reaction was subjected to polyadenylation. A homopolymeric poly(A) tail was added to the 3’ end of the RNA molecules using a Poly(A) Polymerase Tailing Kit (Illumina, USA). Briefly, 1 μg of total RNA was polyadenylated using 2 units of poly(A) polymerase and incubated for 20 min at 37 °C. The poly(A) tailing reaction was stopped by standard phenol:chloroform extraction. Two RT-PCR amplicons, corresponding to the 5’ and 3’ ends, were cloned. Three clones for each cloning reaction were sequenced in both directions by the Sanger method, and the sequence was assembled using Geneious Prime 2021. The mean read depth for the final assembly of CCMoV-M was 20,618X average coverage per nucleotide with 8.52% (3,758,035 reads) of the total reads mapping against CCMoV-M. The complete genome sequence of CCMoV-M (13,800 nt) was submitted to the GenBank database (NCBI) and assigned the accession number MW731536.
Sequence analysis predicted seven ORFs with products arranged in a similar way to those of plant rhabdoviruses, with a 197-nt-long 3’ untranslated (UTR) leader and a 148-nt-long 5’ UTR trailer. Further comparisons of the 3’ and 5’ UTR regions of CCMoV-M revealed that 20 of 32 nucleotides were complementary. This feature had been associated with a putative panhandle structure involved in replication [1]. Interestingly, the first 21 nt of the 3’ leader RNA sequence of CCMoV shared a high level of sequence identity (81%) with the equivalent region in maize fine streak virus (MFSV). A similar level of nucleotide sequence identity (82%) was observed for the last 17 nt of the 5’ trailer RNA sequences of CCMoV-M and MFSV. All of the putative ORFs of CCMoV-M are flanked by gene junctions with the consensus sequence 3’-UUU(A/U)UUUU GUAG UUG-5' (Fig. 1B). This sequence is highly conserved between CCMoV-M and MFSV (Supplementary Fig. S1).
Protein homologs for the predicted proteins encoded by the seven ORFs of CCMoV-M were identified using BLASTp (NCBI). The putative gene order for CCMoV-M is 3’-nucleocapsid (N), phosphoprotein (P), unknown protein (p3), unknown protein (p4), matrix (M), glycoprotein (G), viral polymerase (L)-5’ (Fig. 1C). ORF1 (nt 282-1,667) codes for a putative protein of 50.46 kDa with homology to rhabdovirus nucleoproteins, sharing the highest level of identity (59.27%, aa level) to the nucleoprotein (N) of maize fine streak virus. The putative protein encoded by ORF2 (nt 1,893-2,900) has an estimated size of 37.86 kDa. This protein is homologous to the phosphoprotein (P) of maize fine streak virus, sharing 36.7% identity at the amino acid level. The putative protein encoded by ORF3 (nt 3,166-3,441) has a predicted molecular weight of 10.65 kDa and shares 24% of identity (aa level) with its counterpart P3 in MFSV. ORF4 (nt 3,581-4,579) codes for a protein of 37.05 kDa with 62% identity to its equivalent P4 in MFSV.
The protein encoded by ORF5 (nt 4,754-5,491) has an estimated molecular weight of 28.31 kDa and is homologous to the matrix protein (M) encoded by maize fine streak virus, with 43.70% identity at the amino acid level. ORF 6 (nt 5,795-7,585) codes for a putative protein of 67.39 kDa with homology to rhabdovirus glycoproteins (G). The closest hit was to the MFSV G protein, sharing 49.15% identity at the amino acid level. ORF7 (nt 7,738-13,584) codes for a putative protein of 223.52 kDa with homology to the rhabdovirus polymerase (L). The highest levels of identity observed for the ORF7-encoded protein were 60.93% and 34% to its counterpart in MFSV and potato yellow dwarf virus, respectively. A transmembrane domain was found in the glycoprotein (G) using TMHMM - 2.0 [7]. A search for nuclear localization signals (NLSs) using NLStradamus [8] identified a putative NLS at amino acid position 197 to 207 (KEIAKRIDKIR) in the phosphoprotein (P). The gene order observed in CCMoV-M is analogous to that of MFSV, the only member of the genus Gammanucleorhabdovirus. The amino acid sequence identity values across the coding regions of CCMoV and MFSV were as follows: 59.27% (N), 36.7% (P), 24% (P3), 62% (P4), 43.70% (M), 49.15% (G), 60.93% (L).
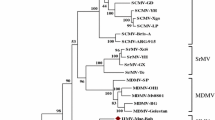
The phylogenetic relationships of CCMoV among members of the subfamily Betarhabdovirinae were estimated from maximum-clade-credibility trees (MCC) generated using the amino acid sequence of the viral polymerase (L) [1]. The full-length polymerase (L) sequences of 38 rhabdoviruses belonging to the subfamily Betarhabdovirinae were retrieved from the ICTV resource page for the family Rhabdoviridae [1]. Sequences were aligned using MAFFT [10]. MCC trees were inferred using BEAST.v1.10.4 [11], and the Whelan and Goldman model of amino acid substitutions, the gamma (G) + invariant (I) sites model of site heterogeneity, and a strict molecular clock (coalescent: constant size) with a random starting tree to perform 10 million Markov chain Monte Carlo runs. The analysis was sampled at every 1000 states. Tree Annotator v1.10.4 was used to output the results of the MCC tree model and calculate posterior probabilities with 10% of the total number of iterations as a burn-in (1 million states). The MCC-inferred tree placed CCMoV in a sister clade with MFSV, the sole member of the genus Gammanucleorhabdovirus (Fig. 2). The placement of CCMoV within the genus Gammanucleorhabdovirus is supported by the MCC-inferred phylogeny and the analogous genome architecture to MFSV. The species demarcation criteria for viruses in the genus Gammanucleorhabdovirus has not yet been established [1]; however, the low levels of identity observed for homologous proteins in MFSV suggest that CCMoV should be considered a distinct member of the genus Gammanucleorhabdovirus. The genome sequence generated in this study can be used for the development of RT-PCR-based detection protocols to investigate the current status of CCMoV in cereal crops.
Maximum-clade-credibility (MCC) consensus tree constructed using amino acid sequences of the full-length viral polymerase (L) of 38 rhabdoviruses belonging to the subfamily Betarhabdovirinae. Branch lengths are drawn to scale, with the scale bar showing the number of substitutions per site. Tip labels are color coded to label different genera within the subfamily Betarhabdovirinae
Data availability
The genomic sequence determined in this study was submitted to the GenBank database under the accession number MW731536.
References
Walker PJ, Freitas-Astúa J, Bejerman N et al (2021) ICTV virus taxonomy profile: Rhabdoviridae. J Gen Virol (in press)
Greber RS (1979) Cereal chlorotic mottle virus—a rhabdovirus of Gramineae in Australia transmitted by Nesoclutha pallida (Evans). Aust J Agric Res 30:433–443. https://doi.org/10.1071/AR9790433
Lockhart BEL (1986) Ocurrence of cereal chlorotic mottle virus in Northern Africa. Plant Dis 70:912–915. https://doi.org/10.1094/PD-70-912
Greber RS, Gowanfock DH (1979) Cereal chlorotic mottle virus purification, serology and electron microscopy in plant and insect tissues. Aust J Biol Sci 32:399–408. https://doi.org/10.1071/BI9790399
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Bushnell B (2022) BBTools software package. https://sourceforge.net/projects/bbmap/
Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Nguyen Ba AN, Pogoutse A, Provart N, Moses AM (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform 10:1–11. https://doi.org/10.1186/1471-2105-10-202
Tsai C, Redinbaugh MG, Willie KJ et al (2005) Complete genome sequence and in planta subcellular localization of maize fine streak virus proteins. J Virol 79:5304–5314. https://doi.org/10.1128/JVI.79.9.5304
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Suchard MA, Lemey P, Baele G et al (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:1–5. https://doi.org/10.1093/ve/vey016
Acknowledgements
Robert A. Alvarez-Quinto is supported by the Republic of Ecuador through the Ecuadorean Science and Technology Secretariat (SENESCYT).
Funding
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project #1009997 and the University of Minnesota, Twin Cities.
Author information
Authors and Affiliations
Contributions
Conceptualization: Benham E. Lockhart, Dimitre Mollov. Formal analysis: Robert A. Alvarez-Quinto, Samuel Grinstead. Investigation: Robert A. Alvarez-Quinto, Benham E. Lockhart, Joana Serrano. Methodology: Benham E. Lockhart, Dimitre Mollov. Supervision: Benham E. Lockhart. Writing—original draft: Robert A. Alvarez-Quinto. Writing—review & editing: Robert A. Alvarez-Quinto, Benham E. Lockhart, Joana Serrano, Samuel Grinstead, Dimitre Mollov. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ralf Georg Dietzgen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

705_2022_5527_MOESM1_ESM.jpg
Supplementary file1 Supplementary Fig. S1 Sequence alignment of the gene junctions detected in cereal chlorotic mottle virus (CCMoV-M) and maize fine streak virus (MFSV). All sequence differences are highlighted in black (JPG 69 KB)
Rights and permissions
About this article
Cite this article
Alvarez-Quinto, R.A., Lockhart, B.E., Serrano, J. et al. Complete genome sequence of a Moroccan isolate of cereal chlorotic mottle virus. Arch Virol 167, 2347–2350 (2022). https://doi.org/10.1007/s00705-022-05527-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05527-2