Abstract
In April 2011, a virus was isolated by single-lesion isolation on Chenopodium quinoa leaves from an amaryllis plant with chlorotic ringspots in a private garden in Changhua County, Taiwan. An Illumina MiSeq sequencing system was used to determine the genomic nucleotide (nt) sequence of the virus. A de novo-assembled contig with 9377 nt, containing an open reading frame encoding a putative potyviral polyprotein, was annotated as the potyvirus Amazon lily mosaic virus (ALiMV), sharing 95.5% nt sequence identity with a partial genomic sequence of ALiMV available in the GenBank database. Therefore, the amaryllis virus was designated as ALiMV-TW. Through 5´ and 3´ rapid amplification of cDNA ends (RACE), the complete 9618-nt genome sequence of ALiMV-TW was determined. Sequence comparisons indicated that the genome and polyprotein of ALiMV-TW share 52.3–65.1% nt and 30.1–64.2% aa sequence identity, respectively, with those of other potyviruses. This is the first report of a complete genome sequence of ALiMV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Amaryllis (Hippeastrum hybridum Hort.) is a perennial bulbous plant of the family Amaryllidaceae that is popular worldwide because of its charming and colorful flowers. The Netherlands, South Africa, Japan, Brazil, and the United States are the world’s major exporters of amaryllis [1], and it is a very popular garden flower in Taiwan. Virus-like symptoms can often be observed on amaryllis plants. The orthotospovirus capsicum chlorosis virus, the cucumovirus cucumber mosaic virus, the potyvirus hippeastrum mosaic virus (HiMV), and the carlavirus nerine latent virus (NeLV) have been reported infecting amaryllis in Taiwan, and mixed infections with HiMV and NeLV are common [2,3,4].
In April 2011, amaryllis plants showing chlorotic ringspots were found in a private garden in Changhua County, Taiwan (Fig. 1a). A virus denoted AV-N1 was isolated from a diseased amaryllis plant by three successive single-lesion transfers on Chenopodium quinoa leaves. AV-N1 induced local chlorotic spots on C. quinoa leaves 7 days post-inoculation (dpi) (Fig. 1b) and caused systemic mosaic and curling symptoms on Nicotiana benthamiana plants at 10 dpi (Fig. 1c). The virus was maintained in C. quinoa as a local-lesion host and in N. benthamiana as a systemic host by mechanical transmission under temperature-controlled conditions (25-28°C) in the greenhouse for study.
Symptoms associated with Amazon lily mosaic virus (ALiMV) infection. (a) Chlorotic ringspots on amaryllis leaves. (b) Chlorotic spots on a Chenopodium quinoa leaf 7 days after inoculation with ALiMV. (c) Systemic mosaic and curling symptoms on a Nicotiana benthamiana plant 10 days after ALiMV infection.
To determine the genomic sequence of AV-N1, 100 mg of AV-N1-infected C. quinoa leaf tissue collected at 7 dpi was used to purify total RNA using a Plant Total RNA Miniprep Purification Kit (GMbiolab, Taichung, Taiwan), following the manufacturer’s instructions. The total RNA was used as a template to prepare a random-primed cDNA library, following the Illumina TruSeq RNA Sample Preparation v2 Guide as described previously [5]. Sequencing was conducted at Genetech Biotech Co., Ltd. (Taipei, Taiwan) using an Illumina MiSeq sequencing platform to synthesize paired-end reads. A total of 18,913,596 reads were obtained after the trimming of adaptors and low-quality sequences. The raw reads were randomly clipped into 23-mers using CLC Genomics Workbench 6.0.2 (CLC bio, Aarhus, Denmark) to facilitate de novo sequence assembly. The contigs were then subjected to Basic Local Alignment Search Tool (BLAST) searches against the non-redundant protein database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). A contig of 9377 nt containing an open reading frame (ORF) encoding a putative potyviral polyprotein was annotated as Amazon lily mosaic virus (ALiMV), a member of the genus Potyvirus, sharing 95.5% nt identity with a partial genomic sequence of ALiMV (accession no. AB158523) available in the GenBank database of NCBI. There were 8,735,130 reads (46.18%) mapped to the ALiMV-annotated contig with minimum and maximum coverage of 11 and 200,890, respectively (Supplementary Fig. S1). Based on sequence similarity, AV-N1 was identified as an amaryllis isolate of ALiMV and renamed ALiMV-TW.
To determine the complete genome sequence of ALiMV-TW, 5ʹ and 3ʹ rapid amplification of cDNA ends (RACE) was performed as described previously [6], using specific primers designed from the nt sequence of the ALiMV-assembled contig. For 5ʹ RACE, the primer ALiMV-P1-1269R (5ʹ-CCGCTATCCCCTTTCCGTAAAGTA-3ʹ) was used to synthesize the first cDNA strand, which was then tailed with PolyC(3c3t11c) (5ʹ-CCCTTTCCCCCCCCCCC-3ʹ) [6]. Subsequently, the primers ALiMV-P1-1137R (5ʹ-GTATGCGTGCTGCTTCTCGATTG-3ʹ) and PolyG(11g3a3g) (5ʹ-GGGGGGGGGGGAAAGGG-3ʹ) [6] were used to amplify the 5ʹ end of the ALiMV genome. The primer ALiMV-NIb-8642F (5ʹ-AACACTGGTACTTCCGGGACGCA-3ʹ) paired with oligo-dT (5´-TTTTTTTTTTTTTTTTTTTTTTTTB-3ʹ) was used for 3ʹ RACE. Reverse transcription (RT) was performed at 50°C for 30 min, followed by inactivation at 94°C for 2 min. Polymerase chain reaction (PCR) was run with the conditions of hot start at 94°C for 2 min, followed by 30 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min and a final extension of 72°C for 7 min. All amplicons were cloned using a TOPO TA Cloning Kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) using standard procedures and sequenced by Mission Biotech (Taipei, Taiwan) using an ABI 3730 DNA sequencer. Three clones were sequenced for each fragment. The nt sequences of the 5ʹ and 3ʹ ends were added to the contig sequence to obtain the complete genome sequence of ALiMV-TW, which was deposited in the GenBank database under accession no. OL355031.
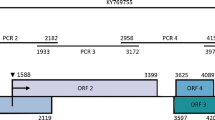
Analysis of the genome structure indicated that the ALiMV-TW genome is 9618 nt long and contains a single ORF, extending from nt 126 to nt 9359 and encoding a polyprotein of 3077 amino acids (aa) (350.8 kDa). According to the consensus sequences for the P1, HC-Pro, and NIa protease cleavage sites [7], the positions of P1 (nt 126-992), HC-Pro (nt 993-2360), P3 (nt 2361-3467), 6K1 (nt 3468-3623), CIP (nt 3624-5525), 6K2 (nt 5526-5681), VPg (nt 5682-6245), NIa (nt 6246-6983), NIb (nt 6984-8540), and CP (nt 8541-9356) were deduced (Fig. 2a). PIPO within P3 (nt 2882-3079, starting from GGAAAAAA) was also found (Fig. 2a).
Genome sequence analysis of Amazon lily mosaic virus (ALiMV). (a) Genome structure of ALiMV. The nucleotide positions corresponding to individual open reading frames are indicated, and putative polyprotein cleavage sites are shown. (b) Color-coded pairwise matrix of potyviral polyproteins. Pairwise Sequence Demarcation Tool Version 1.2 (SDTv1.2) was used to perform pairwise alignments and to generate the color-coded matrix. Comparisons of ALiMV with other potyviruses are highlighted with red boxes. (c) Phylogenetic analysis based on amino acid sequences of the potyvirus polyproteins. The tree was constructed by the neighbor-joining method with 1000 bootstrap replicates using MEGA X software, and the maximum composite likelihood method was used to compute evolutionary distances. The virus names and the accession numbers of genome sequences are shown in Supplementary Table S1. ALiMV is indicated by arrows. Artichoke latent virus (ArLV), belonging to the genus Macluravirus, was used as an outgroup
The complete genome sequences of 136 potyviruses available in the GenBank database (Supplementary Table S1) were used for comparison with the ALiMV-TW sequence. Multiple sequence alignments were performed using the ClustalW method in MEGA X (http://www.megasoftware.net) [8]. Sequence Demarcation Tool Version 1.2 (SDTv1.2) was used to perform pairwise alignments and generate color-coded pairwise matrices and nt and aa sequence identity calculations [9]. Phylogenetic analysis was conducted by the neighbor-joining method with 1000 bootstrap replicates using MEGA X software, and the maximum composite likelihood method was used to compute evolutionary distances. The results revealed that the full-length genome and polyprotein of ALiMV share 52.3%-65.1% nt and 30.1%-64.2% aa sequence identity, respectively, with those of other potyviruses (Fig. 2b). According to the species demarcation criteria of <76% nt and <82% aa sequence identity in the large ORF and its protein product [10], ALiMV is a distinct member of the genus Potyvirus that clusters closely with members of the potato virus Y (PYV) clade (Fig. 2c).
Using a One-Step RT-PCR Kit (GMbiolab), a one-step RT-PCR method with the primer pair ALiMV-P1-967F (5ʹ-CAATAAGACACCTAGTGCCCGAAAGC-3ʹ)/ALiMV-P1-1269R was developed to specifically detect ALiMV in plants. The amplification conditions were RT at 50°C for 30 min, inactivation at 94°C for 2 min, and then 35 cycles of PCR at 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min and a final extension of 72°C for 7 min. A DNA fragment of the expected size (303-bp) was amplified and sequenced directly by Mission Biotech using an ABI 3730 DNA sequencer to verify its correctness.
ALiMV was first discovered in Amazon lily (Eucharis grandiflora) in Japan. Before this report, only part of the genome sequence was available in the GenBank database [11, 12]. Here, we have identified an amaryllis virus as ALiMV and determined its complete genome sequence. Although an ALiMV isolate (accession no. AY590143) was reported previously in Taiwan [13], that virus should be re-identified as HiMV, since it has over 90% nt sequence identity to HiMV isolates. Therefore, this is also the first report of ALiMV in Taiwan.
References
Wang Y, Chen D, He X, Shen J, Xiong M, Wang X, Zhou D, Wei Z (2018) Revealing the complex genetic structure of cultivated amaryllis (Hippeastrum hybridum) using transcriptome-derived microsatellite markers. Sci Rep 13(8):10645
Chen CC, Huang CH, Cheng YH, Chen TC, Yeh SD, Chang CA (2009) First report of Capsicum chlorosis virus infecting amaryllis and blood lily in Taiwan. Plant Dis 93:1346
Chen CC, Chiang FL (2015) Identification and detection of Cucumber mosaic virus isolated from bloody lily (Haemanthus multiflorus). J Taiwan Agric Res 64:74–79
Chen CC, Chang CA, Chiang FL (2016) Serological and molecular identification of Nerine latent virus infecting blood lily (Haemanthus multiflorus Martyn). J Taiwan Agric Res 65:194–206
Chou WC, Lin SS, Yeh SD, Li SL, Peng YC, Fan YH, Chen TC (2017) Characterization of the genome of a phylogenetically distinct tospovirus and its interactions with the local lesion-induced host Chenopodium quinoa by whole-transcriptome analyses. PLoS ONE 12:e0182425
Li JT, Yeh YC, Yeh SD, Raja JAJ, Rajagopalan PA, Liu LY, Chen TC (2011) Complete genomic sequence of watermelon bud necrosis virus. Arch Virol 156:359–362
Goh CJ, Hahn Y (2021) Analysis of proteolytic processing sites in potyvirus polyproteins revealed differential amino acid preferences of NIa-Pro protease in each of seven cleavage sites. PLoS ONE 16:e0245853
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Muhire BM, Varsani A, Martin DP (2014) SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277
Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM, ICTV Report Consortium (2017) ICTV virus taxonomy profile: Potyviridae. J Gen Virol 98:352–354
Terami F, Honda Y, Fukumoto F (1995) Amazon lily mosaic virus, a new potyvirus infecting Amazon lily (Eucharis grandiflora). Ann Phytopathol Soc Jpn 61:1–6
Fuji S, Terami F, Furuya H, Naito H, Fukumoto F (2004) Nucleotide sequence of the coat protein genes of Alstroemeria mosaic virus and Amazon lily mosaic virus, a tentative species of genus Potyvirus. Arch Virol 149:1843–1849
Wu WC, Chang YC (2003) A new mosaic disease of Amazon lily in Taiwan. New Dis Rep 8:29
Funding
This study was funded by the Ministry of Science and Technology, Taiwan, R.O.C. (Grant number MOST 110-2313-B-468-001).
Author information
Authors and Affiliations
Contributions
Funding acquisition: TCC. Supervision: TCC. Investigation: CCL and TCC. Data analysis: CCL and TCC. HTS data analysis: SSL. Manuscript writing: CCL and TCC.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study does not contain any experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ralf Georg Dietzgen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, CC., Lin, SS. & Chen, TC. Complete genome sequence of Amazon lily mosaic virus isolated from amaryllis (Hippeastrum hybridum Hort.). Arch Virol 167, 1495–1498 (2022). https://doi.org/10.1007/s00705-022-05449-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05449-z