Abstract
A novel bipartite begomovirus infecting Cnidoscolus urens (Euphorbiaceae) from Pernambuco State, Brazil, has been characterized. The complete DNA-A (2657 nt) and DNA-B (2622 nt) components of the viral isolates show the typical genome organization of New World bipartite begomoviruses. DNA-A of the isolates had the highest percentage of nucleotide sequence identity (88.6–88.9%) to cnidoscolus mosaic leaf deformation virus. Based on the current classification criteria for the genus Begomovirus, the virus infecting C. urens should be considered a new member of the genus, and the name "cnidoscolus mild mosaic virus" is proposed for the virus, and the name "Begomovirus caboniensis" is proposed for its species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The family Geminiviridae includes viruses with a circular single-stranded DNA genome (2.6–5.2 kb) encapsulated in geminated quasi-icosahedral particles [1]. This family is divided into 14 genera based on their genome organization, type of vector, phylogenetic relationships, host range, and nucleotide sequence similarity [1]. Members of the genus Begomovirus have one or two genomic components known as DNA-A and DNA-B, are transmitted by whiteflies of the Bemisia tabaci complex [2, 3], and infect a wide range of cultivated and non-cultivated plants, mainly in tropical and subtropical regions [4].
Several different begomoviruses have been reported in non-cultivated plants of the family Euphorbiaceae [5,6,7,8,9,10,11]. In Cnidoscolus urens, a wild plant found in northeastern Brazil, only two viruses have been reported: cnidoscolus mosaic leaf deformation virus (CnMLDV) [12] and cnidoscolus blistering yellow mosaic virus (CnBYMV) [13]. Here, the molecular characteristics of a new bipartite begomovirus infecting C. urens in Pernambuco, Brazil, are described.
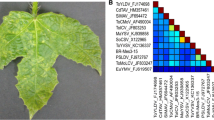
A sample of C. urens showing mild mosaic and leaf deformation symptoms was collected in 2019 in Cabo de Santo Agostinho, Pernambuco State, Brazil (Fig. 1), and total DNA was extracted using the CTAB method [14]. Complete viral genomes were amplified by rolling-circle amplification (RCA) [15], cloned, and sequenced commercially by primer walking (Macrogen Inc., South Korea).
The complete nucleotide sequences of the DNA-A and DNA-B genomic components were assembled using CodonCode Aligner v. 6.0.2 and initially analyzed using the BLASTn algorithm [16] to identify the virus with the highest nucleotide sequence similarity. Pairwise sequence identity comparisons were performed using SDT 1.2 software [17].
Multiple sequence alignments were made using the MUSCLE algorithm implemented in MEGA6 [18]. Phylogenetic analysis was performed on the CIPRES Science Gateway [19] using MrBayes v.3.3.3 [20]. The GTR+I+G model was selected as the best nucleotide substitution model for the DNA-A and DNA-B component datasets. The analysis was carried out using two replicates with four chains each for 10 million generations and sampling every 1000 steps (a total of 10,000 trees). The first 2500 trees were discarded as burn-in. Trees were viewed and edited using Figtree v.1.4
A total of five sequences were obtained: three for the DNA-A component and two for the DNA-B component. The sequence of DNA-A is 2657 nt in length (GenBank accession nos. MZ465527, MZ465586, and MZ465587), and that of DNA-B is 2622 nt in length (MZ465585 and MZ46526). The sequences revealed a typical New World begomovirus gene organization, with DNA-A encoding a coat protein (CP) in the virion-sense strand and a replication-associated protein (Rep), a trans-acting protein (TrAP), a replication enhancer (Ren), AC4, and AC5 in the complementary-sense strand, and with DNA-B encoding two proteins, a nuclear shuttle protein (NSP) in the virion-sense strand and a movement protein (MP) in the complementary-sense strand (Supplementary Table S1). Analysis of the common region (CR) showed a conserved nonanucleotide (5'-TAATATT/AC-3') and three putative iterons (GGGT), one of which was inverted (Supplementary Fig. S1). The CR (200 nt) showed 96% nucleotide sequence identity between the two components, suggesting that they are a cognate pair.
Pairwise comparisons showed that the clones BR_Ca96_19A (MZ465527), BR_Ca112_19A (MZ465586), and BR_Ca113_19A (MZ465587) shared 88.6–88.9% sequence identity with CnMLDV (NC_038982; Supplementary Fig. S2). Based on established criteria for species demarcation within the genus Begomovirus [1], this virus, for which we propose the name "cnidoscolus mild mosaic virus" (CnMMV), should be considered a member of a new species, for which we propose the name "Begomovirus caboniensis", according to the binomial nomenclature system recently established by ICTV [21].
Bayesian inference phylogenetic trees (Fig. 2) were constructed to analyze the DNA-A and DNA-B components. In both trees, the isolates from this study clustered with a CnMLDV isolate (KT966772), confirming their genetic relationship. No evidence of recombination was detected using the RDP 4 program [22].
In the present study, a new bipartite begomovirus infecting C. urens was characterized. Host-range and infectivity studies are needed to confirm the potential of C. urens as a virus reservoir.
References
Zerbini FM et al (2017) ICTV virus taxonomy profle: Geminiviridae. J Gen Virol 98:131–133
Barbosa LF et al (2014) Indigenous American species of the Bemisia tabaci complex are still widespread in the Americas. Pest Manag Sci 70:1440–1445
Navas-Castillo J et al (2011) Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol 49:219–248
Rojas MR et al (2018) World management of Geminiviruses. Annu Rev Phytopathol 56:637–677
Hernandez Z et al (2007) Molecular characterization and experimental host range of Euphobia mosaic virus-Yucatan Peninsula a begomovirus species in the Squash leaf curl virus. Plant Pathol 56:763–770
Hussain K et al (2011) Complete nucleotide sequence of a begomovirus and associated betasatellite infecting croton (Croton bonplandianus) in Pakistan. Arch Virol 156:1101–1105
Fernandes FR et al (2011) Molecular and biological characterization of a new Brazilian begomovirus, euphorbia yellow mosaic virus (EuYMV), infecting Euphorbia heterophylla plants. Arch Virol 156:2063–2069
Fiallo-Olivé E et al (2013) Complete genome sequences of two begomoviruses infecting weeds in Venezuela. Arch Virol 158:277–280
Simmonds-Gordon RN et al (2014) First report of a complete genome sequence for a begomovirus infecting Jatropha gossypifolia in the Americas. Arch Virol 159:2815–2818
Srivastava A et al (2015) Molecular characterization of a new begomovirus associated with leaf yellow mosaic disease of Jatropha curcas in India. Arch Virol 160:1359–1362
Vaca-Vaca JC et al (2018) Croton golden mosaic virus: a new bipartite begomovirus isolated from Croton hirtus in Colombia. Arch Virol 163:3199–3202
Melo AM et al (2016) Cnidoscolus mosaic leaf deformation virus: a novel begomovirus infecting euphorbiaceous plants in Brazil. Arch Virol 161:2605–2608
Oliveira MHC et al (2021) Cnidoscolus blistering yellow mosaic virus: a new begomovirus isolated from Cnidoscolus urens in Brazil. Comunicata Sci 12:1–5
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Inoue-Nagata AK et al (2004) A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J Virol Methods 116:209–211
Altschul SF et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Muhire BM et al (2014) SDT: uma ferramenta de classificação de vírus baseada no alinhamento de sequência de pares e cálculo de identidade. PLoS ONE 9(9):e108277
Tamura K et al (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Miller MA et al (2010) The CIPRES portals. CIPRES
Ronquist F et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Sys Bio Adv 61:539–542
Walker PJ et al (2021) Changes to virus taxonomy and to the International Code of Virus Classifcation and Nomenclature ratifed by the International Committee on Taxonomy of Viruses. Arch Virol 166:2633–2648
Martin DP et al (2015) RDP4 detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Acknowledgments
The authors thank the Universidade Federal de Alagoas. M.M.M.F was the recipient of a CAPES postdoctoral fellowship. L.F.G.C was the recipient of a FAPEAL doctoral fellowship.
Funding
This research was funded by FAPEAL (Fundação de Amparo à Pesquisa do Estado de Alagoas).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chaves, L.F.G., Ferro, M.M.M., de Lima, M.O. et al. Cnidoscolus mild mosaic virus: a new bipartite begomovirus isolated from Cnidoscolus urens in Brazil. Arch Virol 167, 1003–1005 (2022). https://doi.org/10.1007/s00705-022-05367-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05367-0