Abstract
The entomopathogenic fungus Beauveria bassiana is used worldwide for biological control of insects. Seven dsRNA segments were detected in a single B. bassiana strain, RCEF1446. High-throughput sequencing indicated the presence of three mycoviruses in RCEF1446. Two were identified as the known mycoviruses Beauveria bassiana victorivirus 1 and Beauveria bassiana polymycovirus 1, and the novel mycovirus was designated as "Beauveria bassiana bipartite mycovirus 1" (BbBV1). The complete sequence of the BbBV1 is described here. The mycovirus contains two dsRNA segments. The RNA 1 (dsRNA 4) of BbBV1 is 2,026 bp in length, encoding a RNA-dependent RNA polymerase (RdRp) (68.54 kDa), while the RNA 2 (dsRNA 6) is 1,810 bp in length, encoding a hypothetical protein (35.55 kDa) with unknown function. Moreover, the amino acid sequence of RdRp showed the highest sequence identity of 62.31% to Botryosphaeria dothidea bipartite mycovirus 1. Phylogenetic analysis based on RdRp sequences revealed that BbBV1 represents a distinct lineage of unassigned dsRNA mycoviruses infecting fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Mycoviruses (fungal viruses) are pervasive in filamentous fungi, yeasts, and oomycetes [1]. The majority of known mycoviruses have double-stranded RNA (dsRNA) genomes, but mycoviruses with positive-sense (+) single-stranded RNA (ssRNA), negative-sense (-) sRNA, and single-stranded circular DNA (ssDNA) have been reported [2,3,4]. To date, dsRNA mycoviruses are classified into eight families (Amalgaviridae, Chrysoviridae, Hypoviridae, Megabirnaviridae, Partitiviridae, Quadriviridae, Reoviridae, and Totiviridae), and the unassigned genus Botybirnavirus by the International Committee on Taxonomy of Viruses (ICTV) (https://talk.ictvonline.org/). However, many dsRNA mycoviruses have not been assigned to an established family. Among these mycoviruses, some have been grouped into a proposed family or genus, and others have been recognized as members of an unassigned group. A number of bipartite mycoviruses belonging to an unassigned group have been identified in various fungi. Their genomes consist of two double-stranded RNA (dsRNA) segments, but their RdRp sequences show more similarity to the RdRps of proposed unirnaviruses, which have a mono-segment genome, than to those of partitiviruses, which have a two-segment genome. Twenty-three bipartite mycoviruses have been reported so far, including Corynespora cassiicola bipartite mycovirus 1, Botryosphaeria dothidea bipartite mycovirus 1, and Trichoderma harzianum bipartite mycovirus 1 [5,6,7].
The entomopathogenic fungus Beauveria bassiana has a wide host range and is used as a biological control agent for agricultural and forest insect pests [8]. Many B. bassiana strains have been isolated from natural populations, and previous studies have shown that mycoviruses infecting this fungus are widespread and exhibit a high degree of diversity. A number of dsRNA mycoviruses isolated from B. bassiana have been reported previously, including viruses belonging to the families Partitiviridae, Totiviridae, Amalgaviridae, Polymycoviridae, and the proposed family “Unirnaviridae” [9].
Recently, Beauveria bassiana partitivirus 3 (BbPV-3), a member of the proposed genus "Epsilonpartitivirus", was reported in B. bassiana strain RCEF5853 in our laboratory [2]. In this study, we found Beauveria bassiana victorivirus 1, Beauveria bassiana polymycovirus 1, and another novel mycovirus coinfecting B. bassiana strain RCEF1446, which was deposited in the Research Center for Entomogenous Fungi of Anhui Agricultural University (RCEF). Beauveria bassiana victorivirus 1 was the first virus of the genus Victorivirus, family Totiviridae, discovered in B. bassiana, and Beauveria bassiana polymycovirus 1 is a virus causing mild hypervirulence to Galleria mellonella [9]. In this study, the complete genome sequence of another bipartite dsRNA virus and its genome organization were analyzed, and this virus was designated as ‘‘Beauveria bassiana bipartite mycovirus 1’’ (BbBV1).
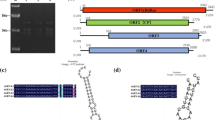
The strain RCEF1446, isolated from Formicidae, was identified as B. bassiana based on its translation elongation factor 1-α gene sequence and morphological characteristics. To investigate the presence of mycoviruses in B. bassiana RCEF1446, the fungus was grown for 5 days on SDAY medium (1% w/v peptone, 4% w/v dextrose, 0.2% w/v yeast extract, and 1.5% w/v agar) overlaid with a sterilized cellophane disc, and dsRNA was extracted from the fresh mycelia using a CF-11 cellulose (Sigma) chromatography method described in a previous report [10]. These dsRNAs were digested with DNase I and S1 nuclease (TaKaRa, Dalian, China). Finally, purified dsRNA was electrophoresed in a 1.5% (w/v) agarose gel and separated into seven distinct bands of approximately 5.2, 2.4, 2.2, 2.0, 1.9, 1.8, and 1.3 kb in length (Fig. 1A). All dsRNAs were sequenced on an Illumina HiSeq 2500 platform at BGI (Shenzhen, China), and the sequences were cleaned up for assembly and analysis. All clean reads were aligned to the fungal genome using bowtie2 (version 2.2.9), and the unaligned reads were captured in a separate file using a flag. A de novo assembly of these unaligned reads was created using Trinity (v2.1.1), and the resultant contigs were analyzed for open reading frames (ORFs) using TransDecoder (version 2.1). In general, when paired-end data were used, the data present within pair one were sufficient for identification of viral contigs; however, for the sake of completeness, following identification of a viral genome, the pipeline was re-run using both pairs. Additionally, a step was added between fastq-dump and bowtie2 in which the reads containing Illumina adaptor sequences were removed using Cutadapt (v1.18). Then, all contigs (>200 nt) were used to search for similar sequences in the GenBank database using BLASTx (http://www.ncbi.nlm.nih.gov/). The results indicated that the fungus was infected by three different mycoviruses. RT-PCR showed that dsRNA 1 (5.2-kb) belonged to Beauveria bassiana victorivirus 1, and dsRNA 2 (2.4-kb), dsRNA 3 (2.2-kb), dsRNA 5 (1.9-kb), and dsRNA 7 (1.3-kb) belonged to Beauveria bassiana polymycovirus 1. However, contig 45 corresponding to dsRNA 4 (2.0 kb) had the highest sequence identity (64.17%) to Botryosphaeria dothidea bipartite mycovirus 1 (QUP79399.1) (Supplementary Fig. S1), while contig 24 corresponding to dsRNA 6 (1.8-kb) had 50.72% amino acid sequence identity to Corynespora cassiicola bipartite mycovirus 1 (QNC69630.1). Thus, dsRNA 4 and dsRNA 6 were segments of a potential novel mycovirus. To obtain the terminal sequences of dsRNA 4 and dsRNA 6, RNA-ligase-mediated rapid amplification of cDNA ends (RLM-RACE) was performed as described by Coutts and Livieratos [11], and the PCR-amplified products were then cloned into pMD18-T (TAKARA) and sequenced separately three times. The complete sequences of dsRNA 4 and dsRNA 6 were assembled using DNAMAN 7.0 (Lynnon Biosoft, USA) and deposited in the GenBank database (accession numbers MW265927 and MW265928). The putative ORFs of both dsRNAs were predicted using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) (Fig. 1B), and the amino acid sequence of the putative RdRp of BbBV1 was aligned with those of other dsRNA viruses using the Multiple Alignment using Fast Fourier Transform (MAFFT) program [12]. A phylogenetic tree was constructed by the maximum-likelihood (ML) method with the LG+G+I+F model and 1000 bootstrap replicates, using MEGA X [13].
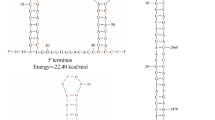
Analysis of the dsRNA profile and genomic organization of the novel mycovirus (BbBV1) from Beauveria bassiana strain RCEF1446. (A) Purified dsRNA extracted from B. bassiana strain RCEF1446 was electrophoresed in a 1.5% agarose gel. M, DNA marker; lane 1, dsRNAs. (B) Schematic representation of the BbBV1 genomic structure. (C) Comparison of the 5′- and 3′-terminal sequences of dsRNA 4 and dsRNA 6 of BbBV1. “*” indicates a conserved nucleotide
The genome of BbBV1 is composed of two segments. The larger segment, named RNA 1, is 2,026 bp in length with a G+C content of 52.8%, while the smaller segment, named RNA 2, is 1,810 bp in length with a G+C content of 55.8%. Each of the segments has only a single ORF in the positive-sense strand. Further analysis indicated that ORF1 (nt 125-1,915) of RNA 1 encodes a putative RNA-dependent RNA polymerase (68.54 kDa), whereas ORF2 (nt 111-1,079) of RNA 2 encodes a protein of unknown function (35.55 kDa). The 5′ UTRs of RNA 1 and RNA 2 were 124 and 110 bp in length, respectively, and further sequence alignment indicated that the 5′ UTRs of BbBV1 RNA 1 and RNA 2 had a highly conserved element (CAUAGAAUUUAAGCCACUGUUCAGCAAACAUU), which is necessary for virus replication [7]. The corresponding 3′ UTRs were 111 and 731 bp in length, and they contained a possible conserved motif despite being quite different in size (Fig. 1C). BLASTp searches demonstrated that the predicted amino acid sequence of ORF1 shared the highest identity of 62.31% (E-value, 0.0; query coverage, 97%) with the RdRp from Botryosphaeria dothidea bipartite mycovirus 1. The amino acid sequence of ORF2 shared the highest identity (50.72%) with a hypothetical protein of unknown function from Corynespora cassiicola bipartite mycovirus 1.
Phylogenetic analysis was performed using the amino acid sequences of RdRps of unclassified viruses related to BbBV1, members of the proposed genera "Ustivirus" [14] and "Unirnavirus" [15], and selected members of the families Partitiviridae and Amalgaviridae. Two members of the family Totiviridae were used as an outgroup [4, 9] (Fig. 2). The phylogenetic tree demonstrated that the unassigned group is much more closely related to members of the proposed family "Unirnaviridae" than to other groups. BbBV1 forms a subclade in the unassigned group with the Curvularia virus 2, Corynespora cassiicola bipartite mycovirus 1, and Podosphaera virus A, and represents a distinct lineage of unassigned dsRNA mycoviruses.
Based on its degree of sequence similarity and phylogenetic clustering with previously reported unassigned dsRNA mycoviruses, the virus BbBV1 from B. bassiana is proposed to be a new member of the unassigned viruses group.
References
Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Shi N, Yang G, Wang P, Wang Y, Yu D, Huang B (2019) Complete genome sequence of a novel partitivirus from the entomogenous fungus Beauveria bassiana in China. Arch Virol 164:3141–3144
Wang Q, Mu F, Xie J, Cheng J, Fu Y, Jiang D (2020) A single ssRNA segment encoding Rdrp is sufficient for replication, infection, and transmission of Ourmia-Like virus in fungi. Front Microbiol 11:379
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA 107:8387–8392
Wang Y, Zhao H, Xue C, Xu C, Geng Y, Zang R, Guo Y, Wu H, Zhang M (2020) Complete genome sequence of a novel mycovirus isolated from the phytopathogenic fungus Corynespora cassiicola in China. Arch Virol 165:2401–2404
Liu H, Wang H, Zhou Q (2021) A novel mycovirus isolated from the plant-pathogenic fungus Botryosphaeria dothidea. Arch Virol 166:1267–1272
Liu C, Li M, Redda ET, Mei J, Zhang J, Elena SF, Wu B, Jiang X (2019) Complete nucleotide sequence of a novel mycovirus from Trichoderma harzianum in China. Arch Virol 164:1213–1216
Huang B, Li CR, Fan MZ, Li ZZ (2002) Moleculer identification of the teleomorph of Beauveria bassiana. Mycotaxon 81:229–236
Kotta-Loizou I, Coutts RHA (2017) Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog 13:e1006183
Okada R, Kiyota E, Moriyama H, Fukuhara T, Natsuaki T (2015) A simple and rapid method to purify viral dsRNA from plant and fungal tissue. J Gen Plant Pathol 81:103–107
Coutts RHA, Livieratos IC (2003) A rapid method for sequencing the 5′- and 3′- termini of double-stranded RNA viral templates using RLM-RACE. J Phytopathol 151:525–527
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Herrero N (2016) A novel monopartite dsRNA virus isolated from the entomopathogenic and nematophagous fungus Purpureocillium lilacinum. Arch Virol 161:3375–3384
Kotta-Loizou I, Sipkova J, Coutts RHA (2015) Identification and sequence determination of a novel double-stranded RNA mycovirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol 160:873–875
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31772226, 31471821 and 30770008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, N., Hu, F., Wang, P. et al. Molecular characterization of two dsRNAs that could correspond to the genome of a new mycovirus that infects the entomopathogenic fungus Beauveria bassiana. Arch Virol 166, 3233–3237 (2021). https://doi.org/10.1007/s00705-021-05239-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05239-z