Abstract
Since late 2018, foot-and-mouth disease (FMD) has reemerged and rapidly swept through pig farms in North and Central Vietnam, despite widespread use of commercial FMD vaccines. To investigate the FMD virus (FMDV) strains responsible for the current epidemics, 40 FMDV samples were collected from 17 provinces during November-December 2018, and the VP1 coding genes were sequenced and analyzed. Phylogenetic analysis and sequence comparisons revealed that all of the reemerging Vietnamese FMDVs belonged to the Mya-98 lineage of the O/Southeast Asia topotype (O/SEA/Mya-98) and shared high nucleotide (99.06–100% identity) and amino acid (97.65–100% identity) sequence similarity with each other. The study results suggested that the reemerging FMDVs originated from local Vietnamese strains. Field viruses had different amino acids in the antigenic sites of VP1 when compared to the strains used in the vaccines. The present study provides an important basis for vaccine selection in the battle against FMD in Vietnam.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foot-and-mouth disease (FMD) is a severe, clinically acute vesicular disease of pigs, cattle, sheep, goats, and many cloven-hoofed wild animals worldwide [3]. FMD is on the World Organization for Animal Health (OIE) list of diseases because of its potential for rapid and extensive spread within and between countries, resulting in severe economic losses [13]. The causative agent of FMD is foot-and-mouth disease virus (FMDV), a member of the genus Aphthovirus in the family Picornaviridae, which is a non‐enveloped, icosahedral virus 26–30 nm in diameter containing a single positive‐sense RNA genome approximately 8,300 nucleotides in length [3]. The genome encodes a single polyprotein that is cleaved into four structural proteins (VP1–VP4) and 10 non-structural proteins (L, 2A–2C, 3A, 3B1–3B3, 3C, and 3D). Sixty copies of the four structural proteins (VP1-4) form the complete viral capsid. The structural proteins VP1–3 are exposed on the surface of the virus, while VP4 is located internally. Among the virus proteins, the VP1 capsid protein is critically involved in virus attachment, protective immunity, and serotype specificity. Various critical antigenic sites and residues of serotype O viruses have been identified on the VP1 protein including aa 144-159 [16], aa 25-41, aa 200-213 [4], and aa 66-80 [7], and residues at positions 43 and 44, 144, 148, 149, 154, 198, and 208 [2, 14, 15]. The sequence of the VP1 gene is used to identify the FMDV lineage and to investigate the genetic relatedness of FMDV isolates [9]. Most of our knowledge about the molecular epidemiology and global distribution of FMDV is based on analysis of the VP1 gene [8].
FMDV is classified into seven distinct serotypes, i.e., O, A, C, Asia 1, Southern African Territories (SAT) 1, SAT 2 and SAT 3 [1]. FMDV infection is endemic in large areas of Africa, Asia, the Middle East, and South America. Seven geographical FMDV pools have been described in endemic areas that share similar viruses. FMDV isolates in eastern Asia and South-East Asia (including Vietnam), where there have been reports of high disease incidence and various serotypes of FMDV throughout the years [5, 18], belong to pool 1 [5]. Since the first record of FMDV in Vietnam in 1956 (as reported on the World Reference Laboratory for Foot-and-Mouth Disease website http://www.wrlfmd.org), serotypes O and A have been endemic, while serotype Asia 1 has not been detected in Vietnam since 2007. In the past two decades, FMDV strains identified as responsible for sporadic epidemics in Vietnam include O/ME-SA/PanAsia, O/SEA/Mya-98, O/CATHAY, and A/SEA/Sea-97 [12, 17]. In May 2015, the O/ME-SA/Ind-2001 lineage virus was detected for the first time in the southern provinces of Vietnam [20].
Due to the endemic circulation and severe losses caused by FMDV in Vietnam, commercial FMD vaccines have been commonly employed in pigs and ruminants throughout the country, and all FMD vaccines were imported from large, well-known companies such as Merial (with manufacturing in France and England), Biogenesis Bago (Argentina), and Lanzhou Bio-Pharmaceutical Factory of China Animals Husbandry Industry (China). Among the FMDV vaccine viruses, O1 Manisa (O/ME-SA/PanAsia lineage) and O 3039 (O/CATHAY topotype) are the most commonly used in Vietnam and have been used on pig farms for preventing the current FMD outbreak. These two vaccine strains were included in 12 out of 16 vaccine products recently recommended for use against FMD by the Department of Animal Health, Ministry of Agriculture and Rural Development of Vietnam [6]. The other recommended products contained O1 Manisa and O Taiwan-98 (O/CATHAY topotype), O1 campos (O/Euro-SA topotype), O/BY/CHA/2010 (O/SEA/Mya-98 lineage), and A22 Iraq and A May 97 (serotype A) strains.
In September 2018, there were several outbreaks in which animals tested positive for FMDV using a VDRG® FMDV 3Diff/PAN Ag Rapid Kit (Median Diagnostics, Korea). FMDV infection has spread rapidly in most of the northern provinces of Vietnam since November 2018. In the FMDV-infected herds, the diseased pigs were reluctant to stand or walk, adopting a dog-like sitting posture, and exhibited depression, loss of appetite, and fever. Severely affected pigs huddled together, had reduced or no food intake, and become lethargic. Vesicles were often observed on the snout and tongue, in the interdigital space at the bulb of the heel, along the coronary band, and on the udder of sows (Fig. 1). Myocarditis lesions characterized by white or grayish stripes or spots were common in animals of various ages that died of the infection (Fig. 1). Pigs of all ages were affected and exhibited degrees of lameness, inappetence, and mortality, which varied by age and immune status. On unvaccinated farms, the morbidity rate sometimes reached 100%, and it was as high as 90% for sucking piglets and 20% for growing pigs. To investigate the FMDV strains responsible for the current large-scale outbreak in Vietnam, the VP1 gene of FMDV isolates collected from pig farms in various affected regions was sequenced and analyzed.
Clinical signs and lesions observed in pigs during recent FMD outbreaks in Vietnam. Grower pigs died from the FMDV infection. (A and B) Vesicles (arrows) on the udders of sows (A) and tongues (B) of FMDV-infected pigs. (C) Sloughed hooves on the front feet of a pig. (D) A myocarditis lesion characterized by white stripes and spots (arrows) in a grower pig
Materials and methods
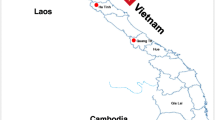
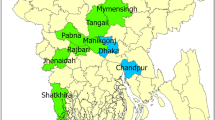
Forty samples of ruptured epidermal tissue were collected from pig farms located in 17 northern and central provinces of Vietnam between November and December 2018 (Table 1 and Fig. 2). Viral RNA was extracted from a 10% specimen homogenate using a QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. cDNA was synthesized using a Superscript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA). A published VN-VP1F/VN-VP1R primer set [11] and an i-MAXTM II DNA Polymerase Kit (iNtRON Biotechnology, Inc., Korea) were used for amplifying the complete VP1 gene using the following thermal profile: 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and a final 10-min extension step at 72°C. The expected size of amplified product was 821 bp. The PCR products were visualized in a 1.2% agarose gel and purified using a QIAquick PCR Purification Kit (QIAGEN, USA).
Geographical locations of FMD outbreaks in the northern and central provinces of Vietnam. The 17 provinces (Bac Giang, Bac Ninh, Dien Bien, Ha Nam, Ha Noi, Ha Tinh, Hai Phong, Hoa Binh, Hung Yen, Lai Chau, Nghe An, Ninh Binh, Phu Tho, Thai Binh, Thai Nguyen, Thanh Hoa, and Vinh Phuc) where the FMDV samples were collected and used in this study are indicated by red dots
RT-PCR primers were used for direct sequencing of the VP1 gene segment using a BigDye Terminator Cycle Sequencing Kit and an automatic DNA sequencer (Model 3730, Applied Biosystems, USA). Nucleotide (nt) and deduced amino acid [7] sequences were edited, assembled, and aligned using Geneious. v9.1.6 software (http://www.geneious.com), and the percent sequence divergence at the nucleotide (nt) and amino acid [7] levels was calculated using the same software. The nt sequences that were obtained were deposited in the GenBank database under accession numbers MN379784-MN379823 as shown in Table 1. Unrooted phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) software, version 7.0.25 [10] by the maximum-likelihood method with 1000 replicates of the bootstrap test. Phylogenetic trees based on the nt sequences of the full-length VP1 gene were generated using the Tamura 3-parameter substitution model with a discrete gamma distribution (TN92 + G).
Results
Molecular diagnostic results revealed that all 40 FMD-suspected farms had at least one FMDV-positive sample. Forty VP1 genes from FMDV isolates from these farms were sequenced. Identical nt sequences were excluded, resulting in the identification of 14 distinct sequences. Phylogenetic analysis based on VP1 gene sequences showed that all of the FMDV strains responsible for the current outbreaks in Vietnam are homogenous and belong to the Mya-98 lineage of the O/Southeast Asia (SEA) topotype (Fig. 3). The viruses formed a monophyletic branch that was most closely related to O/SEA/Mya-98 strains previously identified in Vietnam (O/VN1/2014) and South Korea (O/SKR/2014-JC02D111, O/SKR/2015-JC71GHW, and Jincheon_2014_D1-11). On the basis of BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches using the sequence of the VP1 gene, these reemerging Vietnamese PEDV strains also showed the highest similarity to the Vietnamese strain O/VN1/2014 and South Korean strains collected from outbreaks in 2014-2016. Sequencing results revealed that the VP1 genes of the re-emerging FMDV strains were 639 nucleotides (nt) in length, with 99.06–100% nt and 97.65–100% aa sequence identity to each other. The VP1 genes of these reemerging Vietnamese FMDV strains shared the highest nt sequence identity (94.91‒95.38%) with the Vietnamese strain O/VN1/2014 and 93.9-94.68% identity to the South Korean strains O/SKR/2014-JC02D111, O/SKR/2015-JC71GHW, and Jincheon_2014_D1-11, but only 85.60-86.54% nt sequence identity to most other O/SEA/Mya-98 strains (O/VN/HN2/2014, O/VN/HN3/2014, O/VN/HN5/2014, and O/VN/HN6/2014) identified in Vietnam in 2014. Compared to the serotype O vaccine viruses used in Asian countries, the VP1 genes of the reemerging Vietnamese FMDV strains had the highest (92.18-92.96%) nt sequence identity to those of O/SEA/Mya-98 lineage vaccine viruses isolated recently in China (O/BY/CHA/2010) and South Korea (Yangju-p26, SKR/5/2010, and Andong-p23), but only 75.59-83.57% identity to those of the other the vaccine strains such as O1/Manisa/Turkey/1969, O/TUR/5/2009 Turkey, and O/Lahore/2006 Pakistan, O/HongKong/3039, O/TW/210/1998 Taiwan, O/TUR/5/2009, O1/Campos/1994 Argentina, and O1/Campos/Brazil (Supplementary Material 1).
Phylogenetic tree based on the complete nucleotide sequences of the VP1 coding region of serotype O FMDV isolates. The Vietnamese strains are indicated by solid triangles (▲). Numbers at nodes indicate the level of bootstrap support based on the neighbor-joining analysis of 1000 resampled datasets. Only values above 70% are shown. The scale bar indicates nucleotide substitution per site
To analyze the potential antigenic changes in prevailing FMDV strains, we identified differences in VP1 epitope regions between the vaccine and field strains. Compared to O/SEA/Mya-98 lineage vaccine strains, the Vietnamese field FMDVs have eight amino acid differences (T68A, Q153P, N133S, A137T, S140P, Q153P, A155T, and A198E) at the antigenic sites. Compared to O/ME-SA/PanAsia lineage vaccine strains (O1/Manisa/Turkey/1969, O/TUR/5/2009 Turkey, and O/Lahore/2006 Pakistan) and the O/HongKong/3039 strain, the Vietnamese field strains have 13 different amino acids (Q28H,T68A, N133S, GD137-138TG, TVA140-142PNP, Q153P, A155T, A, T158P, Q198E, and L212S) and 21 (Q28H, A33S, K43T, S68A, S69A,, S74A,, L78V, S134C, GDTSTN137-142TGGPLP, Q153P, AE155-156TA, A158P, T198E, A209V, and L212S) at the antigenic sites (Fig. 4).
Discussion
Since the last quarter of 2018, FMDV infection has reemerged and spread rapidly across northern and central Vietnam. In this study, we discovered that the FMDV strains responsible for the recent, extensive outbreaks belong to the O/SEA/Mya-98 lineage. Viruses of this lineage are normally restricted to mainland Southeast Asia and have been transmitted to and caused devastating outbreaks in five East Asian countries (China, Japan, Mongolia, Russia, and South Korea) since 2010 [19]. These reemerging FMDV strains are most genetically close to a Vietnamese strain identified in 2014 (O/VN1/2014) and are distantly related to the O/SEA/Mya-98 lineage viruses that circulated in other Asian countries prior to 2014. These findings suggest that the reemerging FMDV strains probably originated from local O/SEA/Mya-98 lineage strains that have been evolving and circulating in Vietnam in recent years.
Faced with the spreading FMD epidemic, pigs on most medium- to large-scale farms were immunized with FMD vaccines containing O1 Manisa and O 3039 strains. Antibody prevalence rates achieved four weeks after the second vaccination on many farms generally reached more than 90% (according to recorded veterinary data of the pig-producing companies). However, FMDV infection still reached the farms, indicating that vaccination did not effectively protect swine herds from FMDV infection. The limited protection is possibly the result of heterogeneity between the field strains (0/SEA/Mya-98) and the major vaccine strains (O/ME-SA/PanAsia and O/Cathay). Mismatches in the antigenic regions between vaccine and field FMDV strains were found in sequence comparisons. The presence of various amino acid differences at these antigenic sites implied potential changes in antigenicity, possibly facilitating immune evasion and consequently influencing the efficacy of the vaccines.
To further investigate the antigenic relatedness of vaccine strains to the recently isolated FMDV strains in Vietnam, we collected vaccine matching data for serotype O published by WRLFMD. The data on the website of the laboratory (http://www.wrlfmd.org/east-and-southeast-asia/vietnam#panel-4972) were available only for Vietnamese FMDV isolates collected from 2014 to early 2018 (supplementary material 2). For the field O/SEA/Mya-98 FMDV isolates, the vaccines containing O/HongKong/3039 strain were likely to confer protection against field isolates collected during 2014-2015, but not against isolates collected during 2016-2017. The data also showed that the available vaccines containing the O1/Manisa/Turkey/1969 strain were unlikely to confer protection against O/SEA/Mya-98 strains. This revealed that the major vaccines (containing O/HongKong/3039 and O1/Manisa/Turkey/1969 strains) were probably unable to protect animals from the recent O/SEA/Mya-98 FMDV strains collected during 2014-2017. This suggests a possible similar tendency with respect to vaccine efficacy against the FMDV strains circulating in late 2018, although more investigation is needed to confirm this.
This study provides more insight into the FMDV strains responsible for the extensive outbreaks that occurred in late 2018 in Vietnam. Our findings will provide a useful basis for selection of vaccines in the field. Because it is crucial to antigenically match the vaccine as closely as possible to the causative field isolate, the available vaccines containing O/SEA/Mya-98 FMDV should be optimized for use in the field before further vaccine matching tests can be performed. Our results also suggested that the development of new vaccines containing the circulating O/SEA/Mya-98 FMDV strains may be necessary for vaccination-based control of FMD in the future.
References
Abdela N (2017) Sero-prevalence, risk factors and distribution of foot and mouth disease in Ethiopia. Acta Trop 169:125–132
Aktas S, Samuel AR (2000) Identification of antigenic epitopes on the foot and mouth disease virus isolate O1/Manisa/Turkey/69 using monoclonal antibodies. Rev Sci Tech 19:744–753
Alexandersen S, Mowat N (2005) Foot-and-mouth disease: host range and pathogenesis. Curr Top Microbiol Immunol 288:9–42
Bittle JL, Houghten RA, Alexander H, Shinnick TM, Sutcliffe JG, Lerner RA, Rowlands DJ, Brown F (1982) Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature 298:30–33
Brito BP, Rodriguez LL, Hammond JM, Pinto J, Perez AM (2017) Review of the Global Distribution of Foot-and-Mouth Disease Virus from 2007 to 2014. Transbound Emerg Dis 64:316–332
Department of Animal health MoAaRdoV (2018) Food-and-mouth disease prevalence and instruction in vaccine selection for using in Vietnam. In: Department of Animal health MoAaRdoV (ed) 1635 /TY-DT, Ha noi, Vietnam
Gerner W, Carr BV, Wiesmuller KH, Pfaff E, Saalmuller A, Charleston B (2007) Identification of a novel foot-and-mouth disease virus specific T-cell epitope with immunodominant characteristics in cattle with MHC serotype A31. Vet Res 38:565–572
Knowles NJ, Samuel AR (2003) Molecular epidemiology of foot-and-mouth disease virus. Virus Res 91:65–80
Knowles NJ, Wadsworth J, Bachanek-Bankowska K, King DP (2016) VP1 sequencing protocol for foot and mouth disease virus molecular epidemiology. Rev Sci Tech 35:741–755
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874
Le VP, Lee KN, Nguyen T, Kim SM, Cho IS, Khang DD, Hien NB, Van Quyen D, Park JH (2012) A rapid molecular strategy for early detection and characterization of Vietnamese foot-and-mouth disease virus serotypes O, A, and Asia 1. J Virol Methods 180:1–6
Le VP, Vu TT, Duong HQ, Than VT, Song D (2016) Evolutionary phylodynamics of foot-and-mouth disease virus serotypes O and A circulating in Vietnam. BMC Vet Res 12:269
Leforban Y, Gerbier G (2002) Review of the status of foot and mouth disease and approach to control/eradication in Europe and Central Asia. Rev Sci Tech 21:477–492
Mahapatra M, Aggarwal N, Cox S, Statham RJ, Knowles NJ, Barnett PV, Paton DJ (2008) Evaluation of a monoclonal antibody-based approach for the selection of foot-and-mouth disease (FMD) vaccine strains. Vet Microbiol 126:40–50
Mahapatra M, Parida S (2018) Foot and mouth disease vaccine strain selection: current approaches and future perspectives. Expert Rev Vaccines 17:577–591
Pfaff E, Mussgay M, Bohm HO, Schulz GE, Schaller H (1982) Antibodies against a preselected peptide recognize and neutralize foot and mouth disease virus. EMBO J 1:869–874
Rweyemamu M, Roeder P, Mackay D, Sumption K, Brownlie J, Leforban Y, Valarcher JF, Knowles NJ, Saraiva V (2008) Epidemiological patterns of foot-and-mouth disease worldwide. Transbound Emerg Dis 55:57–72
Sumption K, Rweyemamu M, Wint W (2008) Incidence and distribution of foot-and-mouth disease in Asia, Africa and South America; combining expert opinion, official disease information and livestock populations to assist risk assessment. Transbound Emerg Dis 55:5–13
Valdazo-Gonzalez B, Timina A, Scherbakov A, Abdul-Hamid NF, Knowles NJ, King DP (2013) Multiple introductions of serotype O foot-and-mouth disease viruses into East Asia in 2010-2011. Vet Res 44:76
Vu LT, Long NT, Brito B, Stenfeldt C, Phuong NT, Hoang BH, Pauszek SJ, Hartwig EJ, Smoliga GR, Vu PP, Quang LTV, Hung VV, Tho ND, Dong PV, Minh PQ, Bertram M, Fish IH, Rodriguez LL, Dung DH, Arzt J (2017) First detection of foot-and-mouth disease virus O/Ind-2001d in Vietnam. PLoS One 12:e0177361
Acknowledgments
This research was supported by the Vietnam National University of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. The samples were collected from pigs that died on the farms before sampling. Thus, no aggressive intervention toward the pigs was carried out for sampling purposes.
Additional information
Handling Editor: Tim Skern.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Van Diep, N., Ngoc, T.T.B., Hoa, L.Q. et al. O/SEA/Mya-98 lineage foot-and-mouth disease virus was responsible for an extensive epidemic that occurred in late 2018 in Vietnam. Arch Virol 165, 2487–2493 (2020). https://doi.org/10.1007/s00705-020-04763-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04763-8