Abstract
Porcine reproductive and respiratory syndrome virus 2 (PRRSV2) is a major threat to the global pig industry, particularly in China, the world’s largest pig-rearing and pork-production country. Continuously monitoring the epidemiological and genetic characteristics of PRRSV epidemic strains is beneficial for prevention and control of infection. Previously, we reported the epidemiological and genetic characteristics of PRRSV2 in China from 2012 to 2016. Here, the epidemiological and genetic characteristics of PRRSV2 in China from 2017 to 2018 are reported. During these two years, we collected different types of porcine samples from 2428 pig farms in 27 provinces in China. Of the 7980 samples collected, 2080 (26.07%) were positive for PRRSV2 ORF5 by RT-PCR. The positive rate of PRRSV detection between different regions of China ranged from 8.12% to 29.33%, and from 7.96% to 55.50% between different months. Phylogenetic analysis based on the ORF5 gene revealed that the PRRSV2 strains currently circulating in China belong to five clades, and most of the PRRSVs detected are highly pathogenic PRRSVs (HP-PRRSVs; clade IV) and PRRSV NADC30-like strains (clade I). Sequence analysis revealed multiple amino acid mutation types, including amino acid changes and deletions in both the GP5 and Nsp2 proteins. The presence of these mutations may have an effect on the evolution of the virus by altering the viral titer and/or affecting the antibody response against the virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine reproductive and respiratory syndrome (PRRS), commonly manifested as reproductive failure in sows and respiratory disorders in pigs of all ages, is widespread in almost all pig-rearing countries of the world, representing a major threat to the global pig industry [1]. The etiological agent of PRRS is PRRS virus (PRRSV), which is a single-stranded positive sense RNA virus belonging to the order Nidovirales, family Arteriviridae [2]. PRRSV strains belong to one of two types, PRRSV1 or PRRSV2 [3]. PRRSV1 mainly includes the European type isolates, which are represented by the Lelystad virus (LV) strain, while PRRSV2 mainly includes the North American (NA) type strains, which are represented by VR-2332 [3]. Both PRRSV1 and PRRSV2 possess a 15-kb RNA genome that contains 10 open reading frames (ORFs): ORF1a, ORF1b, ORF2a, ORF2b, ORF3, ORF4, ORF5, ORF5a, ORF6, and ORF7 [4]. ORFs ORF1a and ORF1b encode at least 16 non-structural proteins (nsps) (Nsp1α, Nsp1β, Nsp2, Nsp2N, Nsp2TF, Nsp3-6, Nsp7α, Nsp7β, and Nsp8-12) while ORF2-ORF7 encode structural proteins (GP2a, E(2b), GP3, GP4, GP5, 5a, M, and N) [4]. Within the PRRSV genome, the nsp2 region and the ORF5 region are most variable genes, and they are considered important targets for analyzing the genetic variation and molecular epidemiology of PRRSV [5,6,7,8,9].
The first report of PRRS in China was in 1995 [10], and since then, the pig industry of the world’s largest pig-rearing and pork-consuming country has been accompanied by this disease [6,7,8]. The first strain (CH-1a) of Chinese PRRSV was isolated in 1996 [10], and in the following 10 years, the disease became a common condition for the Chinese pig production [9]. In 2006, an outbreak of highly pathogenic PRRS (HP-PRRS) in China caused approximately 2,000,000 pigs to be infected, of which 400,000 died, resulting in a loss of 120 million Chinese Yuan [11]. PRRSV strains recovered from this outbreak were highly pathogenic and they experienced gradual variation and progressively accumulated genetic changes [9, 12, 13]. After 2006, HP-PRRSV became the main epidemic PRRSV type in mainland China [7, 8]. However, several novel PRRSV strains, such as GM2 and NADC30-like strains, emerged in China due to RNA recombination between Chinese field strains and vaccine strains or foreign viruses after 2012 [14,15,16,17]. The emergence of these novel PRRSVs is a serious concern in China, and may have an influence on the international trade of pigs and related products. Therefore, it is important to monitor the genetic characteristics of PRRSV strains prevalent in China. Since PRRSV2 is mainly responsible for the epidemic in China [6,7,8, 15], we previously monitored and reported the epidemiological and genetic characteristics of PRRSV2 in mainland China from 2012 to 2016 [7, 8]. Here, we report the epidemiological and genetic characteristics of PRRSV2 in mainland China from 2017 to 2018. Our aim is to continuously monitor the epidemiological and genetic characteristics of PRRSV2 in China.
Materials and methods
Sample collection and processing
Between January 2017 and December 2018, a total of 7980 samples, including blood, lungs, spleens, lymph nodes, semen, intestinal tracts, and swine stillbirths were collected from pigs suspected of PRRSV infection on 2057 pig farms in 27 provinces in mainland China (excluding Heilongjiang, Ningxia, Qinghai and Tibet) for PRRSV detection using RT-PCR (Fig. 1). The pigs on many of these farms had been vaccinated with JXA1, Ingelvac®MLV, and/or other commercial vaccines. Blood samples were centrifuged at 4 °C at 5000 rpm for 10 min and the serum was collected, while the tissue samples were homogenized using a Tissue Lyser II (QIAGEN, Germany). Sample homogenates were frozen at −80 °C, thawed three times, and centrifuged at 5000 rpm for 5 min, and the supernatants were then harvested.
A geographic map of China. The numbers of samples positive for PRRSV2 as well as the numbers of total samples collected in different regions of China in 2017 and 2018 are shown. The positive rate for PRRSV2 detection and the detection rates of highly pathogenic PRRSVs (HP-PRRSVs), PRRSV NADC30-like strains, and typical PRRSV strains (C-PRRSVs) in different parts of China are also shown in a table in the lower-left corner of the map. The authors sincerely acknowledge the Ministry of Natural Resources of the People’s Republic of China for providing the map of China free for public use
RT-PCR detection and DNA sequencing
PCR detection was performed as described previously [8]. Total RNA was extracted from the pre-treated samples using an OMEGA Total RNA Kit I (Omega, USA). The PRRSV ORF5 gene and the Nsp2 gene were detected by RT-PCR assays using a PrimeScript One Step RT-PCR Kit Ver.2 (Dye Plus) (TAKARA, Japan) with the RNAs extracted in this study and the primers listed in Table 1. According to a published article [18], the primers for Nsp2 listed in Table 1 are able to amplify a 681-bp product from NADC30-like PRRSV-positive samples, a 984-bp product from HP-PRRSV-positive samples, and a 1074-bp product from typical PRRSV samples. The RT-PCR reaction was performed in a 12.5 μL-volume containing 0.5 μL of PrimeScript One Step Enzyme Mix , 0.5 μL each of the forward primer and the reverse primer, 1.0 μL of template RNA, 4.0 μL of RNase-free water, and 6.0 μL of 2X 1 Step Buffer (Dye Plus). Thermocycler conditions used for PCR were 30 min at 50 °C for reverse transcription, then 5 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 55 °C (for Nsp2) or 60 °C (for ORF5), and 30 s at 72 °C, and a final extension of 10 min at 72 °C. The products were analyzed by 1% ??agarose?? gel electrophoresis. PCR products were purified and cloned into the pMD19-T vector (TAKARA, Japan). Plasmids recovered from the positive colonies were further confirmed by DNA sequencing (Sangon, Shanghai, China). Sequences were submitted to the GenBank database (BioProject number PRJNA587797).
Bioinformatical analysis
For phylogenetic analysis, a Bayesian tree based on the ORF5 gene was generated using BEAST software (version v1.10.0) [19]. Profiles of amino acid changes in Nsp2 and/or GP5 between different PRRSV strains were determined using Clustal X software (version 2.0) [20]. The PRRSV reference strains used in this study and their GenBank accession numbers are listed in Table 2.
Results
RT-PCR detection of PRRSV2 in mainland China from 2017 to 2018
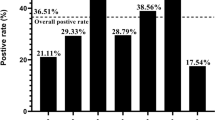
Of the 7980 samples collected, 2080 were found to be positive for the PRRSV ORF5 gene by RT-PCR. The positive rate was 26.07%. The positive rates for PRRSV detection between different parts of mainland China, (Northeast China including Liaoning and Jilin; North China, including Tianjin, Beijing, Shanxi, and Inner Mongolia; East China, including Anhui, Shandong, Jiangsu, Zhejiang, Fujian, Shanghai, and Jiangxi; South China, including Guangdong, Guangxi, and Hainan; Central China, including Hubei, Hunan, and Henan; Southwest China, including Yunnan, Guizhou, Chongqing, and Sichuan; and Northwest China, including Shaanxi, Gansu and Xinjiang) ranged from 8.12% (Northwest China) to 29.33% (South China) (Fig. 1). The positive rates in 2017 and 2018 varied according to the month of sample collection ranging from 7.96% (January) to 55.50% (April) (Fig. 2). The three months with the highest rates of PRRSV detection in 2017 and 2018 were April (55.50%), May (50.35%), and June (38.39%) (Fig. 2).
It has been reported that different types of PRRSV (PRRSV typical strains, HP-PRRSV, and PRRSV NADC30-like trains) can be differentiated according to the size of the PCR product obtained from the nsp2 gene region (1074 bp for typical PRRSV strains, 984 bp for HP-PRRSV strains, and 681 bp for PRRSV NADC30-like strains) [18]. Therefore, we determined the detection rates for the different PRRSV types in China from 2017 to 2018. The results revealed that the proportion of HP-PRRSVs, PRRSV NADC30-like strains, and typical PRRSV strains was 51.57%, 43.30%, and 3.17%, respectively. The detection rate of HP-PRRSV in different regions of China ranged from 33.33% (South China) to 60.11% (Central China) (Fig. 1). The detection rates of typical PRRSV strains in different regions of China ranged from 0 (East China, Southwest China, and Northwest China) to 14.29% (Northeast China) (Fig. 1). The detection rate of PRRSV NADC30-like strains in different regions of China ranged from 38.20% (Central China) to 63.64% (Southwest China) (Fig. 1).
Phylogenetic analysis of PRRSV2
A Bayesian tree generated based on the ORF5 gene revealed that the PRRSV2 strains analyzed in this study belonged to five clades (Fig. 3). Strains in clade I were closely related to the novel American PRRSV2 strain NADC30 [21]. Strains in clade II exhibited a close relationship to GM2, a newly emerged PRRSV2 strain that originated from recombination between the MLV RespPRRS/Repro vaccine strain and the Chinese field strain QYYZ [14]. Strains in clade III and clade V were designated as typical PRRSV2 strains, as strains in these two clades displayed a close relationship to the typical North American PRRSV2 strain VR2332 [22] and the typical Chinese PRRSV2 strain CH-1a [10], respectively. PRRSV2 strains in clade IV were closely related to the HP-PRRSV strain JXA1, which was isolated from the 2006 HP-PRRS outbreak in China [11].
Sequence analysis on the ORF5 gene
Comparisons of ORF5 sequences revealed that the nucleotide sequence identity between the PRRSV strains detected ranged from 61.6% to 100%, and the amino acid sequence identity ranged from 51.8% to 100% (Table 3). The ORF5 regions of strains from North China displayed a low level of nucleotide and amino acid sequence identity to those of strains from East China, and the lowest nucleotide and amino acid sequence identity values were 71.9% and 62.1%, respectively (Table 3). The nucleotide and amino acid sequence identity of ORF5 between the PRRSVs detected in this study and the North American representative strains (VR2332 and NADC30) ranged from 61.9% to 100% and from 68.8% to 100%, respectively (Table 3).
Amino acid sequence alignments revealed that the GP5 proteins of many PRRSVs detected in this study had amino acid changes at aa 11, 29, and 185 compared to that of strain VR2332 (Table 4 and Fig. 4). Compared to the GP5 proteins of the typical PRRSV strains, NADC30-like strains, and GM2-like strains, the GP5 proteins of the HP-PRRSV strains had amino acid changes at aa 9 (G→C), 24 (C→Y), 47 (L→I), and 161 (L→V) (Table 4 and Fig. 4). Compared to the GP5 proteins of HP-PRRSV strains, NADC30-like strains, and GM2-like strains, the GP5 proteins of the typical PRRSV strains had amino acid changes at aa 11 (V→A) and 185 (A→V) (Table 4 and Fig. 4). Compared to the GP5 proteins of typical PRRSV strains, HP-PRRSV strains, and GM2-like strains, the GP5 proteins of the PRRSV NADC30-like strains had amino acid changes at aa 10 (C→Y) and 168 (E→D) (Table 4 and Fig. 4). Compared to the GP5 proteins of GM2-like strains, the GP5 proteins of typical PRRSV strains, HP-PRRSV strains, and NADC30-like strains had an amino acid change at aa 6 (L→S) (Table 4 and Fig. 4). In addition, many PRRSVs detected in some regions had characteristic amino acid changes in the GP5 protein. For example, the amino acid at position 57 of the GP5 proteins of many PRRSVs detected in East China was lysine (K), but the amino acid at the same site of the GP5 proteins of many PRRSVs detected in other parts of China was glutamine (Q) (Table 4).
Sequence analysis on the Nsp2 region
The nucleotide sequence identity values for the Nsp2 region among the PRRSV strains ranged from 39.4% to 100%; and the amino acid sequence identity ranged from 27.9% to 100%. The nucleotide and amino acid sequence identity in Nsp2 between the Chinese PRRSV strains and the North American representative strains (VR2332 and NADC30) ranged from 47.5% to 99.6% and from 26.9% to 98.5%, respectively. Compared to the Nsp2 region of the PRRSV North American type strain VR-2332, the Nsp2 regions of the Chinese HP-PRRSV strains had a discontinuous deletion of 30 amino acids at aa 481 and 533-561 (1+29) (Fig. 5; shown in a green box). The NADC30-like strains detected in this study had a discontinuous deletion of 131 amino acids at aa 322-432, 483, and 504-522 (111+1+19) in the Nsp2 region compared to the Nsp2 region of the typical PRRSV strain VR-2332 (Fig. 5; shown in a red box). Several isolates from this study had other amino acid deletions within Nsp2 compared to that of strain VR2332. For example, the Nsp2 regions of four PRRSV isolates, designated HBEM-1-China-2018-5, HBEM-2-China-2018-5, HBCM-1-China-2018-5, and HBCM-2-China-2018-5, had a discontinuous deletion of 33 amino acids at aa 481, 484-487, and 533-561 (3+1+29), while another four isolates, designated HBJC-1-1-China-2018-5, HBJC-2-China-2018-5, JXNC-China-2018-3, and JX-2-China-2018-3, had a discontinuous deletion of 37 amino acids at aa 481, 533-561, and 612-619 (1+29+7) (Fig. 5).
Discussion
PRRSV1 (previously referred to as the European type) occurs sporadically in China [23], whereas PRRSV2 (previously referred to as the North American type) is more common in China [7, 8, 24,25,26]. Between 2006 and 2016, two large epidemiological studies of PRRSV2 have been conducted in most parts of China [7, 25], and the average positive rate for PRRSV2 in clinical samples was 60.85% from 2006 to 2010 [25] and 55.21% from 2012 to 2015 [7]. In this study, we found that the average positive rate for PRRSV2 in clinical samples in 27 provinces of China was 26.07% from 2017 to 2018. This value is lower than those determined from 2006 to 2010 (60.85%), and from 2012 to 2015 (55.21%), which shows a decreasing trend from 2006 to 2018. In addition, the positive rates in different parts of China from 2017 to 2018 were also considerably lower than those observed from 2012 to 2015 (Northeast China, 10.00% vs. 65.45%; North China, 28.04% vs. 53.35%; East China, 25.06% vs. 63.66%; South China, 29.33% vs. 64.27%; Central China, 27.41% vs. 52.22%; Southwest China, 18.52% vs. 47.70%; Northwest China, 8.12% vs. 47.04%) [7]. These findings suggest that China’s efforts to control PRRS and other swine diseases during the past ten years have had a positive effect. For example, to improve the biosecurity and management level as well as to promote disease control in pig farms, the General Office of the State Council issued the National Medium- and Long-Term Plan on Animal Disease Prevention and Control (2012-2020) in 2012 [27]. This official document establishes goals and priorities for control and eradication of swine diseases in China. In 2016, the Ministry of Agriculture and Rural Affairs issued a national plan for sustainable pig production and development (2016-2020) [28]. This official document aims at promoting the optimization and upgrading of the pig industry. These national actions have contributed to improvements in disease control in the pig industry in China. In addition to the government policy, the outbreak and spread of African swine fever in China has also led to an acceleration in the improvement and enhancement of biosecurity on pig farms in recent years, which may be beneficial for PRRS control in the future.
The history of PRRSV prevalence in China can be divided generally into three stages: the first stage was between 1995 and 2006, in which typical PRRSV strains were the main epidemic type [9], the second stage was between 2006 and 2012, in which HP-PRRSV emerged as the epidemic PRRSV type [25], and the third stage was from 2012 to now. During this stage, HP-PRRSV remains the epidemic type, but novel PRRSV strains such as the NADC30-like strains and GM2-like strains have emerged and are increasing in prevalence [7, 8, 14,15,16, 29]. In the present study, we found that a large proportion of PRRSV strains detected in China from 2017 to 2018 were HP-PRRSV strains (51.57%), while a very low proportion of the detected strains were typical PRRSV strains (3.17%) (Fig. 1). These results are in agreement with the results of the other epidemiological studies conducted in China [6,7,8, 15, 25], suggesting that HP-PRRSV strains are still the epidemic PRRSV type in China. In addition to HP-PRRSVs, NADC30-like strains also represented a large proportion of the total (43.30%). The outbreak of PRRSV NADC30-like strains in China was initially reported on seven intensive pig farms in Beijing, Tianjin, Shanxi, Henan, and Zhejiang in 2014 [29], and the emergence of such strains is likely to have arisen from the recombination between the North American NADC30 strain and the Chinese HP-PRRSV strain [17]. Since then, PRRSV NADC30-like strains have been detected continuously in many regions of China [7, 8, 30,31,32,33]. In this study, the detection rates of PRRSV NADC30-like strains in different regions of China ranged from 38.20% to 63.64%. These values are generally higher than those observed previously in China [7, 8], suggesting that the prevalence of PRRSV NADC30-like strains might be a high-priority problem to be solved in China’s pig industry in the future and should receive more attention.
Considering the unique production model and the lower level of management, vaccination, particularly the use of live vaccines, is the primary choice for majority of pig producers for preventing and controlling PRRS in China [9]. However, the efficacy of inactivated and live vaccines for controlling PRRS in clinical settings has not been as satisfactory as expected. This could explain why many pig farms where the animals had been vaccinated still had high positive rates of PRRSV2 detection in this study. Furthermore, extensive use of live vaccines and the continued spread of HP-PRRSV on pig farms will aggravate the complicated situation of PRRS in China. Selective pressure from the vaccines will increase the genetic diversity of PRRSV epidemic strains, and recombination between field strains and/or between field strains and vaccine strains will accelerate the emergence of novel strains [9]. This complicated situation could be reflected by the observation that there were five clades of PRRSV2 strains including typical strains, HP-PRRSV strains, NADC30-like strains, and GM2-like strains coexisting and spreading together in China, as seen in the phylogenetic analysis in the present study (Fig. 3) as well as other epidemiological studies [8, 23]. These findings suggest that PRRS is a complicated swine disease in China. It is worth noting that the emergence of GM2-like strains (strains in clade II, Fig. 3) is believed to have resulted from recombination between the MLV RespPRRS/Repro vaccine strain and the Chinese field strain QYYZ [14], while the emerging HP-PRRSV strains in China are speculated to be experiencing gradual variation and accumulation of genetic changes, diverging from typical PRRSV viruses [9, 12]. This should be taken into consideration when attempting to develop effective and safe vaccines and choosing vaccines for the prevention and control of PRRS in China [9].
GP5 is an important glycosylated protein of PRRSV, and it is one of the most variable structural proteins among PRRSV strains [34]. Multiple amino acid changes have been found in the GP5 proteins of HP-PRRSVs strains, GM2-like strains, and NADC30-like strains compared to that of typical PRRSV strains, and these changes are commonly seen in its signal peptide coding region (residues 1-26), non-neutralizing epitope (residues 27-30), neutralizing epitope (residues 37-45), and transmembrane regions (residues 66-83, 95-104, and 112-128) [8, 15, 35, 36]. In agreement with these findings, multiple amino acid changes were found in the signal peptide coding region, non-neutralizing epitope, neutralizing epitope, and transmembrane regions of the GP5 proteins of HP-PRRSV strains, GM2-like strains, and NADC30-like strains detected in this study (Fig. 4 and Table 4); these changes may be responsible for the accelerated evolution of this virus [8, 36, 37]. In addition to GP5, the Nsp2 region is also one of the most variable regions among PRRSV strains [8, 34]. Compared to typical PRRSV strains, all of the HP-PRRSV strains detected in this study had a discontinuous deletion of 30 amino acids in their Nsp2 region (Fig. 5; shown in a green box). The presence of this 30-aa deletion in the Nsp2-coding region has been found to be a general characteristic of the HP-PRRSV strains circulation in China [6,7,8, 12, 25]. It is worth noting, however, that this deletion is not related to the virulence of these strains [9, 38]. Nevertheless, since all HP-PRRSV isolates in China have the same 30-aa deletion in their Nsp2-coding region, this deletion can be considered proposed as a genetic hallmark of highly pathogenic PRRSV [9]. The NADC30-like strains that were detected also had a discontinuous deletion of 131 amino acids in their Nsp2 region compared to the Nsp2 region of typical PRRSV strains (Fig. 5; shown in a red box). Similar findings have been reported elsewhere [8, 35]. The presence of these amino acid changes in Nsp2 might affect the antibody response induced by the virus, since amino acid deletions in Nsp2 might alter the viral titer and/or affect the antibody response against PRRSV [39, 40].
In conclusion, we report here the epidemiological and genetic characteristics of PRRSV2 in most parts of China in 2017 and 2018. Our preliminary data revealed that the overall positive rate of PRRSV2 detection in most parts of China showed a decreasing trend compared to that reported between 2006 and 2010 and between 2012 and 2015. Regarding the different types of PRRSV2 detected, HP-PRRSV remains the dominant PRRSV type currently circulating in different regions of China. However, the detection rate of NADC30-like strains is increasing, which should receive more attention. In addition, multiple mutations have been found within the ORF5 region and Nsp2 region, which might accelerate the evolution of this virus and alter the viral titer and/or the antibody response against the virus.
Availability of data and material
Sequences determined in this study were submitted to the GenBank database under the BioProject number PRJNA587797.
References
Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ (2005) Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227(3):385–392
Han M, Yoo D (2014) Engineering the PRRS virus genome: updates and perspectives. Vet Microbiol 174(3–4):279–295. https://doi.org/10.1016/j.vetmic.2014.10.007
Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ (2017) Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol 162(8):2505–2538. https://doi.org/10.1007/s00705-017-3358-5
Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ (2016) Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. https://doi.org/10.1146/annurev-animal-022114-111025
Shi M, Lam TT, Hon CC, Murtaugh MP, Davies PR, Hui RK, Li J, Wong LT, Yip CW, Jiang JW, Leung FC (2010) Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol 84(17):8700–8711. https://doi.org/10.1128/jvi.02551-09
Xie J, Cui T, Cui J, Chen Y, Zhang M, Zhou P, Deng S, Su S, Zhang G (2014) Epidemiological and evolutionary characteristics of the PRRSV in Southern China from 2010 to 2013. Microb Pathog 75:7–15. https://doi.org/10.1016/j.micpath.2014.08.001
Peng Z, Zhao T, Liang W, Song W, Gao Z, Tang X, Chen H, Wu B (2017) RT-PCR detection of porcine reproductive and respiratory syndrome virus based on the ORF5 gene in mainland China, 2012–2015. Acta Virol 61(3):336–340. https://doi.org/10.4149/av_2017_312
Liang W, Zhao T, Peng Z, Sun Y, Stratton CW, Zhou D, Tang X, Tian Y, Chen H, Wu B (2019) Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus circulating in central and South China in 2016. Acta Trop 190:83–91. https://doi.org/10.1016/j.actatropica.2018.11.004
Zhou L, Yang H (2010) Porcine reproductive and respiratory syndrome in China. Virus Res 154(1–2):31–37. https://doi.org/10.1016/j.virusres.2010.07.016
Guo B, Chen Z, Liu W, Cui Y (1996) Isolation and identification of procine reproductive and respiratory syndrome virus from aborted fetues suspected of PRRS. Chin J Anim Poult Infect Dis 26:1–5
Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF (2007) Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2(6):e526. https://doi.org/10.1371/journal.pone.0000526
Zhou L, Chen S, Zhang J, Zeng J, Guo X, Ge X, Zhang D, Yang H (2009) Molecular variation analysis of porcine reproductive and respiratory syndrome virus in China. Virus Res 145(1):97–105. https://doi.org/10.1016/j.virusres.2009.06.014
An TQ, Tian ZJ, Xiao Y, Li R, Peng JM, Wei TC, Zhang Y, Zhou YJ, Tong GZ (2010) Origin of highly pathogenic porcine reproductive and respiratory syndrome virus. China. Emerg Infect Dis 16(2):365–367. https://doi.org/10.3201/eid1602.090005
Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, Baoli S, Yingzuo B (2012) Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol 86(17):9543. https://doi.org/10.1128/jvi.01341-12
Xie J, Zhu W, Chen Y, Wei C, Zhou P, Zhang M, Huang Z, Sun L, Su S, Zhang G (2013) Molecular epidemiology of PRRSV in South China from 2007 to 2011 based on the genetic analysis of ORF5. Microb Pathog 63:30–36. https://doi.org/10.1016/j.micpath.2013.05.013
Lu WH, Tun HM, Sun BL, Mo J, Zhou QF, Deng YX, Xie QM, Bi YZ, Leung FC, Ma JY (2015) Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol 175(2–4):332–340. https://doi.org/10.1016/j.vetmic.2014.11.016
Zhang Q, Jiang P, Song Z, Lv L, Li L, Bai J (2016) Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Vet Microbiol 197:93–101. https://doi.org/10.1016/j.vetmic.2016.11.010
Li C, Zhuang J, Wang J, Han L, Sun Z, Xiao Y, Ji G, Li Y, Tan F, Li X, Tian K (2016) Outbreak Investigation of NADC30-Like PRRSV in South-East China. Transbound Emerg Dis 63(5):474–479. https://doi.org/10.1111/tbed.12530
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4(1):vey016. https://doi.org/10.1093/ve/vey016
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Brockmeier SL, Loving CL, Vorwald AC, Kehrli ME Jr, Baker RB, Nicholson TL, Lager KM, Miller LC, Faaberg KS (2012) Genomic sequence and virulence comparison of four Type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res 169(1):212–221. https://doi.org/10.1016/j.virusres.2012.07.030
Halbur PG, Paul PS, Meng XJ, Lum MA, Andrews JJ, Rathje JA (1996) Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J Vet Diagn Invest 8(1):11–20. https://doi.org/10.1177/104063879600800103
Liu J, Wei C, Lin Z, Fan J, Xia W, Dai A, Yang X (2019) Recombination in lineage 1, 3, 5 and 8 of porcine reproductive and respiratory syndrome viruses in China. Infect Genet Evol 68:119–126. https://doi.org/10.1016/j.meegid.2018.12.006
Guo Z, Chen XX, Li X, Qiao S, Deng R, Zhang G (2019) Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central China during 2016-2017: NADC30-like PRRSVs are predominant. Microb Pathog 135:103657. https://doi.org/10.1016/j.micpath.2019.103657
Li B, Fang L, Guo X, Gao J, Song T, Bi J, He K, Chen H, Xiao S (2011) Epidemiology and evolutionary characteristics of the porcine reproductive and respiratory syndrome virus in China between 2006 and 2010. J Clin Microbiol 49(9):3175–3183. https://doi.org/10.1128/jcm.00234-11
Guo Z, Chen XX, Li R, Qiao S, Zhang G (2018) The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J 15(1):2. https://doi.org/10.1186/s12985-017-0910-6
Council GOotS (2012) The national medium and long-term plan on animal disease prevention and control (2012–2020). http://www.gov.cn/gongbao/content/2012/content_2152427.htm
China TMoAaRAotPsRo (2016) National plan for sustainable pig production and development (2016–2020).http://www.moa.gov.cn/nybgb/2016/diwuqi/201711/t20171127_25920859.htm
Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H (2015) NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis 21(12):2256–2257. https://doi.org/10.3201/eid2112.150360
Wang LJ, Xie W, Chen XX, Qiao S, Zhao M, Gu Y, Zhao BL, Zhang G (2017) Molecular epidemiology of porcine reproductive and respiratory syndrome virus in Central China since 2014: The prevalence of NADC30-like PRRSVs. Microb Pathog 109:20–28. https://doi.org/10.1016/j.micpath.2017.05.021
Zhang H, Leng C, Ding Y, Zhai H, Li Z, Xiang L, Zhang W, Liu C, Li M, Chen J, Bai Y, Kan Y, Yao L, Peng J, Wang Q, Tang YD, An T, Cai X, Tian Z, Tong G (2019) Characterization of newly emerged NADC30-like strains of porcine reproductive and respiratory syndrome virus in China. Arch Virol 164(2):401–411. https://doi.org/10.1007/s00705-018-4080-7
Zhou L, Kang R, Yu J, Xie B, Chen C, Li X, Xie J, Ye Y, Xiao L, Zhang J, Yang X, Wang H (2018) Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-Like, Jxa1-Like, and Mlv-Like Strains. Viruses. https://doi.org/10.3390/v10100551
Zhou L, Kang R, Xie B, Tian Y, Wu X, Lv X, Yang X, Wang H (2018) Identification of a novel recombinant type 2 porcine reproductive and respiratory syndrome virus in China. Viruses. https://doi.org/10.3390/v10040151
Yu X, Chen N, Wang L, Wu J, Zhou Z, Ni J, Li X, Zhai X, Shi J, Tian K (2012) New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet Microbiol 158(3–4):291–299. https://doi.org/10.1016/j.vetmic.2012.02.036
Zhao K, Ye C, Chang XB, Jiang CG, Wang SJ, Cai XH, Tong GZ, Tian ZJ, Shi M, An TQ (2015) Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol 89(20):10712–10716. https://doi.org/10.1128/jvi.01446-15
Delputte PL, Vanderheijden N, Nauwynck HJ, Pensaert MB (2002) Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J Virol 76(9):4312–4320. https://doi.org/10.1128/jvi.76.9.4312-4320.2002
Music N, Gagnon CA (2010) The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev 11(2):135–163. https://doi.org/10.1017/s1466252310000034
Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, Guo X, Ge X, Yang H (2009) The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol 83(10):5156–5167. https://doi.org/10.1128/jvi.02678-08
Han J, Liu G, Wang Y, Faaberg KS (2007) Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J Virol 81(18):9878–9890. https://doi.org/10.1128/jvi.00562-07
Wang X, Sun L, Li Y, Lin T, Gao F, Yao H, He K, Tong G, Wei Z, Yuan S (2013) Development of a differentiable virus via a spontaneous deletion in the nsp2 region associated with cell adaptation of porcine reproductive and respiratory syndrome virus. Virus Res 171(1):150–160. https://doi.org/10.1016/j.virusres.2012.11.006
Acknowledgements
The authors sincerely acknowledge the Ministry of Natural Resources of the People’s Republic of China for providing the map of China free for public use.
Funding
This study was funded by the National Key Research & Development Program of China (grant number 2018YFD0500800), the Agricultural Science and Technology Innovation Program of Hubei Province (grant number 2018skjcx05), Guangdong Provincial Key Laboratory of Livestock Disease Prevention (grant number YDWS1903), and the earmarked fund for China Agriculture Research System (grant number CARS-35). Wan Liang and Zhong Peng were supported in part by China Postdoctoral Science Foundation (grant numbers: 2019M652609 to W.L. and 2018 M640719 to Z.P.).
Author information
Authors and Affiliations
Contributions
Conceptualization, Z.P. and B.W.; methodology, S.X., W.L., X.W., H.C., J.F., W.S., L.H., X.T.; writing—original draft preparation, S.X., W.L., Z.P.; writing—review and editing, Z.P., H.C., B.W.; supervision, Z.P., B.W.; project administration, Z.P., B.W.; funding acquisition, W.L., Z.P., B.W. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable. All of the clinical samples used in this study were submitted by veterinarians/or the farm owners to the Huazhong Agricultural University Veterinary Diagnostic Laboratory for routine testing.
Additional information
Handling Editor: Sheela Ramamoorthy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xie, S., Liang, W., Wang, X. et al. Epidemiological and genetic characteristics of porcine reproduction and respiratory syndrome virus 2 in mainland China, 2017–2018. Arch Virol 165, 1621–1632 (2020). https://doi.org/10.1007/s00705-020-04661-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04661-z