Abstract
A new cyanophage, S-B05, infecting a phycoerythrin-enriched (PE-type) Synechococcus strain was isolated by the liquid infection method, and its morphology and genetic features were examined. Phylogenetic analysis and morphological observation confirmed that S-B05 belongs to the family Myoviridae of the order Caudovirales. Its genome was fully sequenced, and found to be 208,857 bp in length with a G + C content of 39.9%. It contained 280 potential open reading frames and 123 conserved domains. Ninety-eight functional genes responsible for cyanophage structuring and packaging, DNA replication and regulation, and photosynthesis were identified, as well as genes encoding 172 hypothetical proteins. The genome of S-B05 is most similar to that of Prochlorococcus phage P-TIM68. Homologues of open reading frames of S-B05 can be found in various marine environments, as revealed by comparison of the S-B05 genome sequence to sequences in marine viral metagenomic databases. The presence of auxiliary metabolic genes (AMGs) related to photosynthesis, carbon metabolism, and phosphorus assimilation, as well as the phylogenetic relationships based on AMGs and the complete genome sequence, reflect the phage-host interaction mechanism or the specific adaptation strategy of the host to environmental conditions. The genome sequence information reported here will provide an important basis for further study of the adaptive evolution and ecological role of cyanophages and their hosts in the marine environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacteria, formerly called "blue-green algae", are simple, primitive photosynthetic microorganisms with a widespread occurrence in aquatic ecosystems. So far, 40 different genera of cyanobacteria have been recognized [1]. Unicellular cyanobacteria of the genus Synechococcus are ubiquitously distributed in the euphotic layer of the ocean, where they make a large contribution to the global primary production [2, 3]. Members of the genus Synechococcus are both phenotypically and phylogenetically diverse, which is reflected in the ecogeographic and temporal distribution of different ecotypes [4, 5]. The population size and distribution are also affected by infection with cyanophages (i.e., viruses that infect cyanobacteria), which are known to be active, diverse and abundant in all aquatic environments [6, 7]. It has been suggested that cyanophage infection of Synechococcus is very common and is responsible for the mortality of a significant proportion of the Synechococcus population [8], regulating both its abundance and diversity. However, compared with the large number of different cyanophages in the natural environment, the number of Synechococcus cyanophages that have been isolated and sequenced is still low due to the limitations of traditional microbial isolation and culture technology, which have limited our understanding of cyanophage diversity and ecology. By 2019, 318 Synechococcus cyanomyoviruses, nine Synechococcus cyanopodoviruses, and 13 Synechococcus cyanosiphoviruses had been isolated and identified, with their complete genome sequences published in the NCBI database (Table S1). Some of these genome sequences with the same phage name exhibit sequence identity of 98% or more, including 59 sequences of Synechococcus phage S-RIM2 [9], 45 of Synechococcus phage ACG-2014d, 41 of Synechococcus phage ACG -2014f, 24 of Synechococcus phage ACG-2014a, and 18 of Synechococcus phage ACG-2014b [10] (Table S1).
In this study, to better understand the biological characteristics of marine cyanophages and to explore the potential functional links between cyanophage and cyanobacteria, a novel cyanophage S-B05 infecting an estuarine Synechococcus strain belonging to subcluster 5.1 clade IX was isolated. Its morphological features were examined, and its complete genome sequence was determined, providing insights into the manner in which marine cyanophages adapt to their environment and the potential functional links between cyanophages and cyanobacteria in the marine ecosystem.
Materials and methods
Sampling and cyanophage isolation
The cyanophage S-B05 was isolated from surface seawater at a coastal site (B05, 36°59.894′N, 122°52.542′E) in the Bohai Sea. Fifteen liters of seawater was collected and sequentially filtered through 3-μm (Isopore™ 3.0 μm TSTP; Merck, Ireland) and 0.2-μm (Isopore™ 0.2 μm GTTP; Merck, Ireland) polycarbonate membranes. The filtrates were then concentrated by tangential flow filtration with a 50-kDa cartridge (Pellicon® XL Cassette, Biomax® 50 kDa; polyethersulfone, Millipore Corporation, Billerica, MA, USA) to a final volume of 50 ml and stored at 4℃ [11]. The host of the cyanophage is Synechococcus sp. strain MW02, which belongs to subcluster 5.1, clade IX, and can survive in low-salinity waters [4]. It was grown in 250-ml conical flasks under a constant illumination of approximately 25 µMol photons m−2 s−1 at 25℃ in f/2 seawater medium. Liquid infection was used to isolate the cyanophage [12]. The virus-enriched seawater was added to an exponentially growing Synechococcus culture for adsorption, after which the phage-host suspension was cultured under constant irradiation of 25 µMol photons m−2 s−1 at 25℃. The host culture without addition of the viral concentrate was used as a control. Typically, lysis of the host cells was observed within one week. Putative cyanophage lysates were filtered through a 0.22-μm-pore-size membrane (Millex®-GP 0.22 μm PES; Merck, Ireland), and the filtrate was stored at 4℃ in the dark for further tests [13].
Viruses were purified by serial dilution (maximum probable number [MPN]). Dilution series (tenfold dilutions over seven orders of magnitude) of the lysate were screened for infectivity. The most dilute sample causing lysis was saved, and the process was repeated three times. After the third round, the most dilute sample producing lysis was assumed to contain a single particle [12]. The cyanophage was then concentrated using an Amicon® Ultra 15 device with a 30-kDa ultra-PL membrane (Merck, Ireland) [14]. Further purification was performed by sucrose density gradient centrifugation. Aliquots of viral concentrates were layered on 20-to-50% (w/v) sucrose density gradients and centrifuged at 110,000 × g for 2.5 h at 4℃ in a Beckman Coulter ultra-high-speed centrifuge, using the corresponding polypropylene centrifuge tubes (REF:326,819). The phage was collected using a syringe with a needle, diluted at least 1:5 with buffer STE, and centrifuged at 110,000 × g for 3 h at 4℃. The purified phage precipitates were resuspended in SM buffer and stored at 4℃ [15]. Eight Synechococcus strains, including Synechococcus WH7803, WH8102, MW03, LTWRed, LTWGreen, PSHK05, CCMP1333, and PCC70022, were used to test the host specificity of cyanophage S-B05.
Morphological examination by transmission electron microscopy
Purified viral particles in a volume of 20 μl were placed on a 200-mesh copper grid, which was left for 10 min before the excess sample was removed using filter paper. The grid was stained by adding a drop of 1% (w/v) phosphotungstic acid (pH 7.0), and the excess stain was removed immediately using filter paper [13]. The purified cyanophage particles were then examined using a transmission electron microscope (TEM) (JEOLJEM-1200EX, Japan) at 100 kV to determine the structural characteristics and dimensions [16].
DNA isolation and sequencing
DNA was extracted from sucrose-density-gradient purified phage particles using a TIANamp Virus DNA Kit (TIANGEN). Purified S-B05 genomic DNA was sequenced using Illumina Miseq 2 × 300 paired-end sequencing in an ABI 3730 automated DNA sequencer. Gaps between contigs were closed using GapCloser and GapFiller, with purified genomic DNA as the template [17].
Bioinformatics analysis
The open reading frames (ORFs) in the cyanophage S-B05 genome were predicted using GeneMarkS (https://topaz.gatech.edu/GeneMark/genemarks.cgi), GLIMMER (https://ccb.jhu.edu/software/glimmer/index.shtml) and RAST (Rapid Annotation using Subsystem Technology) (https://rast.nmpdr.org). The predicted ORFs were translated into amino acid sequences, and BLASTp (https://blast.ncbi.nlm.nih.gov/) was used to search for homologous genes in the NCBI non-redundant protein database. The functions of the proteins encoded by the homologous genes were predicted, and the conservation of the amino acid sequences was analyzed as described previously [18]. Protein domains were predicted and analyzed using InterPro (https://www.ebi.ac.uk/interpro/) and CDD (https://www.ncbi.nlm.nih.gov/cdd). tRNA scan-SE 1.21 software was used to search for tRNA genes [19], and RNAmmer v1.2 was used to predict ribosomal RNA in the full genome sequence. Genome mapping was performed using DNAplotter (version 17.0.1).
Comparative analysis of cyanophage genomes
The genome sequence of S-B05 was compared with other completely sequenced phage genomes using tBLASTx. A "proteomic tree" based on genome-wide similarities was generated using viptree (https://www.genome.jp/viptree/) [20]. To choose the phage sequences to be compared, all viral sequences in Virus-Host DB (https://www.genome.jp/virushostdb/), including viruses with complete genome sequences available in NCBI/RefSeq and GenBank whose accession numbers are listed in EBI Genomes, were used to establish a circular tree. Then the 35 closest phages in the circular tree were selected together with phage S-B05 to establish a rectangular tree for subsequent comparison and analysis. Phylogenetic analysis and gene comparison with other related phages were carried out in the ClustalW program using the amino acid sequences of 31 representative auxiliary metabolic genes (AMGs). A maximum-likelihood phylogenetic tree was constructed using MEGA (Version 7.0.18) [21]. Bootstrap values were determined based on 1000 replicates. Average nucleotide sequence identity values were obtained using OrthoANI (Average Nucleotide Identity by Orthology), which used the orthogonal method to determine the overall similarity between the two genomic sequences [22].
Acquisition of reads from metagenome datasets
Sequences of homologues of S-B05 ORFs were obtained from the Global Ocean Sampling (GOS) metagenomic database (0.1–0.8 μm) [23] and Pacific Ocean Virome (POV) database [24]. The GOS database contains 48 metagenomes covering a wide range of distinct surface marine environments and a few estuarine environments, while the POV database contains 13 viral metagenomes sampling at 10 m from four biogeographic regions in the Pacific Ocean that vary by season and proximity to land. The sequences from the two databases were downloaded from the iMicrobe website (https://data.imicrobe.us/). Homologues with E-values < 10–5, alignment values > 30 and score > 40 were selected [25]. The proportions of reads in the metagenomes homologous to S-B05 ORF were calculated by dividing number of homologous reads by the total number of metagenomic reads. Prior to calculating each ratio, repeated reads were deleted. The coverage of each metagenome against the S-B05 ORFs was calculated by dividing the number of homologous ORFs in each metagenome by the number of ORFs in S-B05 [25, 26].
Results
Host range and phage morphology
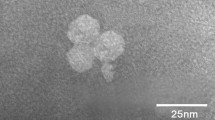
The host of cyanophage S-B05 is PE-type Synechococcus sp. strain MW02 (NCBI accession number KP113680), which was originally isolated from Hong Kong estuarine waters. Cyanophage S-B05 did not infect the other tested Synechococcus strains, indicating its host specificity. Examination by transmission electron microscopy showed that it has an icosahedral head with a diameter of 74 nm and a contractile tail of 152 nm in length (Fig. 1).
General genomic features and tRNA information
The genome of cyanophage S-B05 is a 208,857-bp DNA molecule with a G + C content of 39.9%. It has 280 potential open reading frames, and 123 conserved domains (Table S2). Using the program tRNAscan-SE, 10 different tRNA types were identified in the genome (Arg, Asn, Val, Leu, Ala, Tyr, Ser, Ile, Thr and Pro) (Table 1). The genome of cyanophage S-B05 is slightly larger than those of most members of the family Myoviridae, whose length is usually between 160 and 200 kb with a G + C content of 35%-42%. Functional annotation of predicted ORFs showed that only 98 (36.3%) were associated with specific functions (E-value < 10−5), while the remaining 172 (63.7%) were predicted to encode hypothetical proteins due to insufficient information about homologous genes [27]. The predicted ORFs could be divided into six functional groups related to structure (26 ORFs), packaging (3 ORFs), DNA replication and regulation (32 ORFs), photosystem proteins (6 ORFs), hypothetical proteins, and additional functions related to physiological activity (31 ORFs) (Fig. 2).
Phylogenetic position of cyanophage S-B05
Phylogenetic analysis showed that S-B05 clustered with Prochlorococcus phage P-TIM68, Synechococcus phage S-SM2, and Synechococcus phage Bellamy (Fig. 3b). To compare the genome similarity between S-B05 and other related cyanophages, the average nucleotide sequence identity values were calculated for the nine cyanophages that are most closely related to S-B05 in the phylogenetic tree (Fig. 3c) [14]. The average nucleotide identity values (OrthoANI values) between S-B05 and the other nine phages were all very similar, ranging from 65.11% to 69.55%.
Cyanobacterial-assisted metabolic gene AMGs
A total of 14 AMGs were identified in the genome of cyanophage S-B05, including genes involved in photosynthesis (ORF 234 psbA, ORF236 psbD, ORF 134 hli, ORF277 speD, ORF219 petE, and ORF132 petF), carbon metabolism (ORF141 talC), phosphate acquisition (ORF250 phoH, and ORF135 pstS), DNA biosynthesis (ORF80 cobS, ORF109 mazG, and ORF11 purM), heat shock (ORF269 hsp) and cAMP phosphodiesterase activity (ORF273) (Fig. 4). Cyanophages for comparison were selected based on their phylogenetic position in the whole-genome-based tree (Fig. 3b). A maximum-likelihood phylogenetic tree of the selected cyanophages based on 31 representative AMGs showed that S-B05 did not cluster together with most of the phages (Fig. 4).
A heat map showing the gene copy number matrix of 31 auxiliary metabolic genes (AMGs) of 36 cyanophages. These phages were selected based on their phylogenetic position in the whole-genome-based tree. A maximum-likelihood phylogenetic tree of these cyanophages based on 31 AMGs is shown to the left of the heat map. The names of the cyanophages are colored separately according to the genus of the host from which the phage was isolated: Synechococcus, blue; Prochlorococcus, red
Distribution of S-B05 ORF homologues
Homologues of S-B05 ORFs were found in different marine environments. Of the reads obtained from the POV, the proportion of S-B05 ORF homologues was similar in coastal (1.69%) and open ocean (1.62%) areas and higher in intermediate (2.56%) regions (Table 2). The distribution of reads obtained from the 0.1- to 0.8-μm fraction in the GOS metagenomes showed a similar pattern (Table 2) in which S-B05 ORF homologues had a similar proportion in coastal (1.59%) and open ocean (1.689%) areas. It is noteworthy that the coverage of S-B05 ORFs in the metagenomic databases was very high. Overall, homologues of 97% of the S-B05 ORFs were found in metagenomic databases. In the GOS metagenome database, the coverage of the S-B05 ORFs reached 95% in coastal areas. Even when the data from the POV database were limited to open ocean areas, the lowest coverage still reached 73.7%.
Accession number
A complete genomic sequence of phage S-B05 was submitted to the GenBank database and assigned the accession number MK799832.
Discussion
A linear double-stranded DNA cyanophage, S-B05, was isolated from Bohai Sea seawater in summer. This strain belongs to the family Myoviridae, order Caudovirales, based on the phylogenetic analysis and morphological features observed by transmission electron microscopy. A tRNAscan-SE analysis demonstrated the presence of 10 different tRNA types in the genome (Arg, Asn, Val, Leu, Ala, Tyr, Ser, Ile, Thr and Pro) (Table 1). Cyanophage tRNAs play an important role in phage infection of the host. Currently, only a few freshwater cyanophages have been reported to contain tRNA genes corresponding to all 20 amino acids, such as S-CRM01 [28] and S-CBWM1 [25]. There are also phages that lack only a few tRNA types, for example, the Synechococcus phage S-PM2 genome lacks only tRNA genes for Cys and Phe [29]. Cyanophages infecting marine Synechococcus have not been reported to contain more tRNA types than reported here. In fact, the list of amino acids represented by the tRNAs is very similar in most known viral genomes, suggesting that the tRNA genes may originate from the same gene pool, losing or acquiring genes and generating sequence differences during evolution [25]. Studies have shown that cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus (low G + C content) and Synechococcus (high G + C content) hosts. Some phages in the family Myoviridae that can infect both Prochlorococcus and Synechococcus might overcome the limitations associated with differences in G + C content by carrying an additional set of tRNAs in their genome for AT-rich codons [30].
A comparison of genome sequences revealed that cyanophage S-B05 shares many similar genes with other cyanophages, including Prochlorococcus phage P-TIM68, Synechococcus phage Bellamy, Synechococcus phage S-SM2, and Synechococcus phage S-T4 (Table 3). Interestingly, although the host of S-B05 is Synechococcus, it shares the most genes with cyanophage P-TIM68, whose host is Prochlorococcus. Therefore, it is speculated that phages infecting cyanobacteria may share many same genes. It has been reported that some cyanophages isolated from a member of one genus are able to infect members of another genus, such as Syn19, which is known to infect members of both Synechococcus and Prochlorococcus [31]. Cyanophage S-B05 and P-TIM68 are closely related in terms of their complete genome but distant in terms of OrthoANI, which had a value of only 68%. This may suggest that cyanophages that infect marine cyanobacteria originally had a common ancestor but subsequently diverged.
Viruses can regulate host metabolism and facilitate production of new viruses by providing virus-encoded AMGs. They are commonly found in the genomes of cyanophages [32] and are usually homologues of host genes related to photosynthesis [33, 34], carbon metabolism [35], nucleic acid synthesis and metabolism [36, 37], and stress tolerance [38], providing a genetic basis for evading host immune defenses and improving their ecological adaptability [39, 40]. During the interaction between phages and cyanobacteria, gene transfer promotes the co-evolution between the virus and the host and has an important impact on their genetic diversity [41].
In S-B05, the photosynthesis-related proteins module contains six ORFs. The photosynthetic genes psbA and psbD encode the D1 and D2 proteins of the photosystem II reaction center and are key genes for host light regulation [42]. They are commonly found in the genome of cyanophages isolated along the coast or in the open ocean. Among the 36 viral genomes analyzed, 97% had psbA and 75% had psbD (Fig. 4). Their sequences are conserved and highly similar to the host photosynthetic genes. In fact, the psbA and psbD genes from the phage and the host are homologous [42, 43]. It has been reported that there is a bidirectional exchange of genetic information between the phage and the host genome, and it is thought that the photosynthetic genes of cyanophages were acquired in the process of infection of their hosts [44]. The presence or absence of psbA in a phage genome may be related to the length of the latent period of infection. Whether a phage also carries psbD may reflect constraints on coupling of viral- and host-encoded PsbA–PsbD in the photosynthetic reaction center across divergent hosts [45]. The phylogenetic tree constructed based on psbA genes included both Synechococcus and cyanophage sequences (Fig. 5). However, the host genes clustered in one group while the cyanophage genes clustered in another. This indicates that even though the photosynthetic system genes of the cyanophage are derived from the host gene pool, they still differ from the photosynthetic genes of the host. It has been reported that genes have been transferred from host to phage in a discrete number of events over the course of evolution, followed by horizontal and vertical transfer between cyanophages [45]. The polyamine biosynthesis gene speD, which was found in cyanophage S-B05, is known to catalyze the terminal step in polyamine synthesis in other prokaryotes, and polyamines affect the structure and oxygen evolution rate of the photosystem II (PSII) reaction center in higher plants [46]. Therefore, it may play a role in maintaining the host PSII reaction center during phage infection [47]. Cyanophage S-B05 also contains two photosynthetic electron transport genes coding for plastocyanin (petE) and ferredoxin (petF). Infectious cyanophages carrying photosynthetic system genes not only enhance the photosynthetic ability of the host but also increase the adaptability of cyanophages themselves by offering optimal condition for virus production [45].
S-B05 includes three related genes, cobS, mazG and purM, that are involved in DNA biosynthesis. MazG regulates cell nutrition and stress and is a regulator of programmed cell death [48]. It is a highly conserved gene in cyanopodoviruses and cyanomyoviruses that infect Synechococcus. It is also widespread in marine ecosystems, reflecting the rich diversity of cyanophages. Meanwhile, the cyanophage mazG gene has a small effective population size, indicative of rapid lateral gene transfer. However, a study has shown that the mazG genes in Prochlorococcus and Synechococcus phages do not cluster with the host mazG gene, suggesting that their primary hosts are not the source of the mazG gene [48]. CobS encodes a subunit of an enzyme that is involved in the synthesis of bacterial vitamin B12, an important cofactor for the cyanobacterial ribonucleotide reductase, and plays an important role in the biosynthesis of proteins [47]. DNA synthesis genes such as those encoding purine synthase (purS, purN, purM, purH) and pyrimidine synthase (pyrE) are also usually found in cyanophage genomes. These cyanobacterial enzymes catalyze important stages of DNA biosynthesis to provide nucleosides for phage genome replication and DNA synthesis. However, cyanophage S-B05 contains only the purine operon purM. As shown in Fig. 4, some Prochlorococcus phages, such as P-SSM2, P-SSM7 and P-TIM68, contain several purine and pyrimidine synthase (purS, purM, purN, purH, pyrE), while Synechococcus phages contain only one purine synthase gene (purM or purS), with the exception of S-CAM4, which has both purM and purS. Because purine and pyrimidine syntheses are involved in biosynthesis, the reason for this difference may be related to the growth of the host as well as the growth of the virus itself. Cyanophages that lack purine and pyrimidine synthase, such as S-B05, are presumably dependent on the host for these activities.
Genes related to the regulation of phosphate, phoH and pstS, were found in the genome of S-B05. The pstS gene plays an important role in the phosphate transport system of the host. It can regulate phosphate absorption and assimilation under phosphate-limiting conditions and may contribute to the pyrolysis cycle by increasing the phosphate supply in the host cell [31, 49]. As the most common phosphate-regulating unit gene in the genome of cyanophages, phoH is present in 35 of 36 related cyanophages, with P-TIM68 being the exception (Fig. 4). However, its specific function is still unclear [50]. In this study, the host of S-B05, Synechococcus sp. strain MW02, was isolated from an estuarine site and has been confirmed to belong to subcluster 5.1 clade IX, which includes strains that can survive in low-salinity waters [4]. Since the isolation site is largely influenced by the Pearl River discharge, where low phosphorus levels are common [51], phoH and pstS may play an important role in regulating phosphate uptake in the host in this ecosystem.
The phylogenetic relationships observed based on AMG content are quite different from those based on the complete genome sequence. This indicates that even cyanophages with close kinship have very different AMGs (Fig. 4). For example, in trees based on the AMG genes, S-B05 belongs to a separate branch and is distantly related to Bellamy and S-SM2, which, however, are closely related to S-B05 when the analysis is done based on the whole genome sequence. This could be attributed to various adaptive advantages related to the habitat type of the host [33].
In conclusion, S-B05, a cyanophage belonging to the family Myoviridae, was isolated from a coastal station in the Bohai Sea. Genome sequencing showed that S-B05 has a linear dsDNA genome of 208,857 bp with 280 putative open reading frames. Phylogenetic analysis and morphological observation showed that cyanophage S-B05 belongs to the family Myoviridae, order Caudovirales according to the ICTV virus classification system. Different phylogenetic relationships were observed depending on whether AMGs or whole genome sequences were analyzed, reflecting different phage-host interaction mechanisms or a specific adaptation strategy of the host to environmental conditions. Comparisons with sequences in marine viral metagenomic databases demonstrated that homologues of S-B05 ORFs are present in different marine environments. Because of the limited gene and protein information in the cyanophage database, it is essential to isolate and identify more cyanophages from different environments. The resulting information will provide an important basis for further research of the interaction, adaptive evolution, and ecological role of cyanophages and their hosts in marine environments.
References
Carmichael W (1994) The toxins of cyanobacteria. Sci Am 270(1):78–86. https://www.jstor.org/stable/24942554
Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, Karl DM, Li WKW, Lomas MW, Veneziano D, Vera CS, Vrugt JA, Martiny AC (2013) Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. PNAS 110(24):9824–9829. https://doi.org/10.1073/pnas.1307701110
Buitenhuis ET, Li WKW, Vaulot D, Lomas MW, Landry MR, Partensky F, Karl DM, Ulloa O, Campbell L, Jacquet S, Lantoine F, Chavez F, Macias D, Gosselin M, McManus GB (2012) Picophytoplankton biomass distribution in the global ocean. Earth Syst Sci Data 4(1):37–46. https://doi.org/10.5194/essd-4-37-2012
Xia X, Vidyarathna NK, Palenik B, Lee P, Liu H (2015) Comparison of the seasonal variations of Synechococcus Assemblage structures in estuarine waters and coastal waters of Hong Kong. Appl Environ Microbiol 81(21):7644. https://doi.org/10.1128/AEM.01895-15
Haverkamp T, Schouten D, Doeleman M et al (2009) Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the baltic sea. ISME J 3(4):397. https://doi.org/10.1038/ismej.2008.118
Suttle C, Chan A, Cottrell M (1990) Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467–469. https://doi.org/10.1038/347467a0
Suttle C (2000) Cyanophages and their role in the ecology of cyanobacteria. In: Whitton B, Potts M (eds) The ecology of cyanobacteria. Springer, Dordrecht, pp 563–589. https://doi.org/10.1007/0-306-46855-7_20
Suttle C (1994) Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol 60(9):3167–3174
Marston MF, Martiny JB (2016) Genomic diversification of marine cyanophages into stable ecotypes. Environ Microbiol 18(11):4240–4253. https://doi.org/10.1111/1462-2920.13556
Gregory A, Solonenko S, Ignacio-Espinoza J et al (2016) Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer. BMC Genom. https://doi.org/10.1186/s12864-016-3286-x
Yang Q, Gao C, Jiang Y, Wang M, Zhou X, Shao H, Gong Z, McMinn A (2019) Metagenomic characterization of the viral community of the South Scotia Ridge. Viruses 11(2):95. https://doi.org/10.3390/v11020095
Wilhelm S, Carberry M, Eldridge M et al (2006) Marine and freshwater cyanophages in a Laurentian Great Lake: evidence from infectivity assays and molecular analyses of g20 genes. Appl Environ Microbiol 72(7):4957–4963. https://doi.org/10.1128/AEM.00349-06
Wilson W, Joint I, Carr N et al (1993) Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. Strain WH7803. Appl Environ Microbiol 59(11):3736
Caroline C, Sandra K, Lauro FM (2018) Complete genome sequence of the cyanophage S-PRM1 isolated from Singapore coastal waters. Mar Genom 43:58–60. https://doi.org/10.1016/j.margen.2018.08.005
Vidaver AK, Koski RK, Etten JLV (1973) Bacteriophage φ6: a lipid-containing virus of pseudomonas phaseolicola. J Virol 11(5):799–805
Deveau H, Labrie SJ, Chopin MC et al (2006) Biodiversity and classification of lactococcal phages. Appl Environ Microbiol 72(6):4338–4346. https://doi.org/10.1128/AEM.02517-05
You S, Wang M, Jiang Y et al (2019) The genome sequence of a novel cyanophage S-B64 from the Yellow Sea, China. Curr Microbiol 76:681–686. https://doi.org/10.1007/s00284-019-01680-1
Buttimer C, Born Y, Lucid A, Loessner MJ, Fieseler L, Coffey A (2018) Erwinia amylovora phage vB_EamM_Y3 represents another lineage of hairy Myoviridae. Res Microbiol 169(9):505–514. https://doi.org/10.1016/j.resmic.2018.04.006
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964. https://doi.org/10.1093/nar/25.5.955
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380. https://doi.org/10.1093/bioinformatics/btx157
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100. https://doi.org/10.1099/ijsem.0.000760
Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al (2007) The sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. https://doi.org/10.1371/journal.pbio.0050077
Hurwitz BL, Sullivan MB (2013) The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE 8:e57355. https://doi.org/10.1371/journal.pone.0057355
Xu Y, Zhang R, Wang N, Cai L, Tong Y, Sun Q, Chen F, Jiao N (2018) Novel phage–host interactions and evolution as revealed by a cyanomyovirus isolated from an estuarine environment. Environ Microbiol 20:2974–2989. https://doi.org/10.1111/1462-2920.14326
Lu L, Cai L, Jiao N et al (2017) Isolation and characterization of the first phage infecting ecologically important marine bacteria Erythrobacter. Virol J. https://doi.org/10.1186/s12985-017-0773-x
Liu Z, Wang M, Meng X et al (2017) Isolation and genome sequencing of a novel Pseudoalteromonas phage PH1. Curr Microbiol 74(2):1–7. https://doi.org/10.1007/s00284-016-1175-9
Dreher TW, Brown N, Bozarth CS, Schwartz AD, Riscoe E, Thrash C et al (2011) A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages. Environ Microbiol 13(7):1858–1874. https://doi.org/10.1111/j.1462-2920.2011.02502.x
Mann NH, Clokie MRJ, Millard A et al (2005) The genome of S-PM2, a "Photosynthetic" T4-Type bacteriophage that infects marine Synechococcus strains. J Bacteriol 187(9):3188–3200. https://doi.org/10.1128/JB.187.9.3188-3200.2005
Enav H, Béjà O, Mandel-Gutfreund Y (2012) Cyanophage tRNAs may have a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J 6:619–628. https://doi.org/10.1038/ismej.2011.146
Sullivan MB, Huang KH, Ignacio-Espinoza JC et al (2010) Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol 12(11):3035–3056. https://doi.org/10.1111/j.1462-2920.2010.02280.x
Crummett LT, Puxty RJ, Weihe C (2016) The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 499:219–229. https://doi.org/10.1016/j.virol.2016.09.016
Mann N, Cook A, Millard A, Bailey S, Clokie M (2003) Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741–741. https://doi.org/10.1038/424741a
Philosof A, Battchikova N, Aro E, Beja O (2011) Marine cyanophages: tinkering with the electron transport chain. ISME J 5:1568–1570. https://doi.org/10.1038/ismej.2011.43
Thompson L, Zeng Q, Kelly L, Huang K, Singer A, Stubbe J, Chisholm S (2011) Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. PNAS 108(39):E757–E764. https://doi.org/10.1073/pnas.1102164108
Dwivedi B, Xue B, Lundin D, Edwards R, Breitbart M (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biolss. https://doi.org/10.1186/1471-2148-13-33
Hagay E, Mandel-Gutfreund Y, Béjà O (2014) Comparative metagenomics analyses reveal viral-induced shifts of host metabolism towards nucleotide biosysnthesis. Microbiome. https://doi.org/10.1186/2049-2618-2-9
Kelly L, Ding H, Huang K, Osburne M, Chisholm S (2013) Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J 7:1827–1841. https://doi.org/10.1038/ismej.2013.58
Millard AD, Zwirglmaier K, Downey MJ et al (2009) Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol 11(9):2370–2387. https://doi.org/10.1111/j.1462-2920.2009.01966.x
Huang S, Wang K, Jiao N (2012) Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage–host genetic exchanges. Environ Microbiol 14(2):540–558. https://doi.org/10.1111/j.1462-2920.2011.02667.x
Gao E, Ning D (2014) Advances in researches on cyanophage auxiliary metabolic genes. Microbiol China 41(8):1667–1674. https://doi.org/10.13344/j.microbiol.china.130650
Lindell D, Jaffe JD, Johnson ZI et al (2005) Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438(7064):86–89. https://doi.org/10.1038/nature04111
Lindell D, Sullivan MB, Johnson ZI et al (2004) Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci 101(30):11013–11018. https://doi.org/10.1073/pnas.0401526101
Zeidner G, Bielawski JP, Shmoish M et al (2010) Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ Microbiol 7(10):1505–1513. https://doi.org/10.1111/j.1462-2920.2005.00833.x
Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW (2006) Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. https://doi.org/10.1371/journal.pbio.0040234
Bograh A, Gingras Y, Tajmir-Riahi HA et al (1997) The effects of spermine and spermidine on the structure of photosystem II proteins in relation to inhibition of electron transport. FEBS Lett 402(1):41–44. https://doi.org/10.1016/S0014-5793(96)01453-6
Sullivan MB, Coleman ML, Weigele P et al (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3(5):e144. https://doi.org/10.1371/journal.pbio.0030144
Bryan MJ, Burroughs NJ, Spence EM et al (2008) Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS ONE 3(4):e2048. https://doi.org/10.1371/journal.pone.0002048
Ignacio-Espinoza JC, Sullivan MB (2012) Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes. Environ Microbiol 14(8):2113–2126. https://doi.org/10.1111/j.1462-2920.2012.02704.x
Li X, Sun Y, Wang X, Liu J, Wang G (2017) Research progress of new biomarker gene of phoH for bacteriophage genetic diversity. Biotechnol Bull 33(10):40–45. https://doi.org/10.13560/j.cnki.biotech.bull.1985.2017-0725
Xu J, Glibert PM, Liu H et al (2012) Nitrogen sources and rates of phytoplankton uptake in different regions of Hong Kong waters in summer. Estuaries Coasts 35(2):559–571. https://doi.org/10.1007/s12237-011-9456-9
Acknowledgements
This work was financially supported by the Natural Science Foundation of China (No. 41906126; 41976117; 41676178), and the Open Research Fund of LMB, SCSIO (LMB20091001). We sincerely thank Dr. Xia X. from SCSIO for isolating and providing the Synechococcus culture. We also thank the captain and crew of R/V Dongfanghong II, as well as the students and staff on board the cruise.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, T., Guo, C., Wang, M. et al. Isolation and complete genome sequence of a novel cyanophage, S-B05, infecting an estuarine Synechococcus strain: insights into environmental adaptation. Arch Virol 165, 1397–1407 (2020). https://doi.org/10.1007/s00705-020-04595-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04595-6