Abstract
A total of 472 samples from domestic pigs collected in China from 2015 to 2018 were tested for the presence of porcine circovirus types 2 and 3 (PCV2 and PCV3, respectively) by conventional polymerase chain reaction analysis. The prevalence of PCV2, PCV3, and PCV2/3 co-infection was 50.0%, 13.3%, and 6.78%, respectively. The complete genomic sequences of 66 PCV2 isolates and four PCV3 isolates were determined. Based phylogenetic analysis, the PCV2 isolates were assigned to three genotypes, PCV2a, PCV2b, and PCV2d, representing 13.6% (9/66), 25.8% (17/66), and 60.6% (40/66) of the total, respectively. All four PCV3 isolates shared a high degree of similarity in their complete nucleotide sequences (98.8–99.8% identity) and ORF2 amino acid sequences (98.6–99.5% identity). These results indicate that all three PCV2 genotypes (PCV2a, PCV2b, and PCV2d) are present on pig farms and that PCV2d has become the predominant genotype. The predicted amino acid sequences of the four PCV3 isolates indicated that PCV3-CN-JL53/PCV3-CN-LN56, PCV3-CN-HLJ3, and PCV3-CN-0710, belonged to the genotypes PCV3a, PCV3b, and PCV3a-IM, respectively. In view of the great harm that PCV2 causes to the pig industry, the epidemic trend of PCV3 should continue to be closely monitored. This study provides information about the prevalence, genetic diversity, and molecular epidemiology of PCV2 and PCV3 in China from 2015 to 2018.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine circovirus (PCV), a member of the genus Circovirus, family Circoviridae, has two genotypes: porcine circovirus type 1 (PCV1) and porcine circovirus type 2 (PCV2) [26]. PCV1 was first identified as a contaminant of PK-15 cell culture in 1974 but is generally recognized as non-pathogenic to pigs [34]. PCV2 is the etiological agent of PCV-associated disease (PCVAD), is generally considered to be one of the primary causative agents threatening the swine industry worldwide, and has resulted in large economic losses [22]. Recently, a novel PCV strain, designated as PCV3, was identified by metagenomic sequencing as the cause of acute porcine dermatitis and nephropathy syndrome (PDNS)-like clinical signs and reproductive failure in sows [24]. Moreover, a PCV3 virus stock from the rescue of an infectious PCV3 DNA clone was shown to successfully reproduce PDNS-like disease by PCV3 infection [14].
The PCV genome consists of a closed circular single strand of DNA. Within the genus Circovirus, non-pathogenic PCV1 and pathogenic PCV2 have a similar genomic organization. In most strains, the PCV2 genome is nine nucleotides longer than that of PCV1 (1768 vs. 1759 nucleotides [nt], respectively). PCV2, which is one of the smallest animal viruses, has no envelope and a closed single-stranded DNA genome of 1766–1769 nt [20]. The genome contains four well-characterized open reading frames (ORFs). PCV2 ORF1 encodes two replication proteins (Rep and Rep’) that are both essential for viral replication via the rolling-circle replication mechanism [3]. PCV2 ORF2 encodes the capsid (Cap) protein, which is the only structural component of the virion and the dominant immunogenic antigen [21]. PCV2 ORF3 encodes an apoptotic protein [19], while ORF4 is not essential for viral replication, but has been associated with modulating the host immune response via regulation of CD4+ and CD8+ T lymphocytes during infection [13]. The PCV3 genome has two main ORFs: ORF1, which encodes a protein involved viral replication, and ORF2, which encodes the major structural Cap protein.
Point mutations and genetic recombination contribute to the evolution of PCV2 resulting in the generation of genetic diversity and driving rapid and global spread of the virus. Genotypic shifts may be asociated with differences in pathogenicity and vaccine immunity. Based on pairwise sequence comparisons and linearized phylogenetic trees, as proposed by the EU consortium on PCV disease in 2008, PCV2 is classified into three genotypes: PCV2a, PCV2b, and PCV2c [32]. The PCV2d genotype was first identified by Guo et al. in 2010 [9], and it became the predominant genotype worldwide around 2012 [36]. Meanwhile, a new genotype, designated as PCV2e, was identified in swine in midwestern U.S. states and Mexico from 2006 to 2015 [5, 11]. In addition, a new genotype, designated as PCV2f, was identified in a retrospective study of PCV2 infection in China [1]. With an increasing number of complete PCV2 sequences available, a phylogenetic method based on a p-distance of 0.0035 for ORF2 and 0.02 for the complete sequences was recently proposed for classification of new isolates, but further refinement is needed (www.PCVD.eu). Hence, new criteria have been proposed: a maximum intra-genotype p-distance of 13% (based on the ORF2 gene), bootstrap support at the corresponding internal node of > 70%, and the availability of at least 15 sequences [7]. PCV3 was first reported in the USA in 2016 and has since been detected in China [2], Brazil [35], Korea [15], Japan [10], and many European countries [4, 6, 33]. PCV3 infection has been implicated as the cause of PDNS and reproductive failure, as well as cardiac and multisystemic inflammation [25]. Moreover, increasing evidence has shown that PCV3 coinfection with other pathogens might be associated with increased pathogenicity in pigs [2].
Since 2016, coinfection of PCV3 with PCV2 has frequently been reported in clinical samples from diseased pigs. Zhang et al. reported that, according to the results of a nano-dPCR assay, 14.7% of the samples were positive for PCV2, 16.6% were positive for PCV3, and 6.8% were positive for both viruses [40]. Ku et al. demonstrated that the positive rates for PCV2 and PCV3 in these samples were 62.2% (138/222) and 34.7% (77/222), respectively. The coinfection rate of PCV3 with PCV2 was 15.8% (35/222) [16].
The aim of this study was to analyze the genetic diversity of PCV2 and PCV3 by phylogenetic analysis based on complete genome sequences to provide information regarding the occurrence of PCV2 and PCV3 in pigs in Northeast China from 2015 to 2018.
Materials and methods
Cells and monoclonal antibodies (mAbs) against PCV2
PK-15 cells, free of PCV1 contamination, were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen Corporation, Carlsbad, CA, USA), 100 IU of penicillin per mL, and 100 μg of streptomycin per mL at 37 °C in a humidified 5% CO2 incubator. Based on the PCV2a Cap sequence, 13 mAbs (1H10, 2C1, 3A5, 4D6, 4F5, 5B9, 6B10, 8E10, 8G12, 10D3, 8E4, 5B8, and 1D2) were generated and preserved in our laboratory.
Sample information
A total of 472 samples (including serum, lungs, and lymph nodes) were collected from diseased and healthy pigs in China from 2015 to 2018. Of the 472 clinical samples, 286 were from the northeastern provinces of Liao Ning, Ji Lin, and Hei Longjiang, while the others were collected from the provinces of Shan Dong (30), He Nan (24), Inner Mongolia (32), He Bei (37), Jiang Xi (33), Shan Xi (12), and Guang Xi (18). All samples were stored at -20 °C and submitted to the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (Harbin, China) for detection of PCV2 and PCV3.
Polymerase chain reaction (PCR) analysis of PCV2 and PCV3
Viral genomic DNA was extracted from 200 μL of serum or tissue homogenate using a TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions and stored at -80 °C. The primers used for detection of PCV2 and PCV3 are listed in Table 1. For detection of PCV2 and PCV3, each 25-μL reaction mixture contained 12.5 μL of 2× Taq polymerase premix (TaKaRa Biotech, Dalian, China), 0.25 μL of each primer at a concentration of 10 μM, 2 μL of DNA template, and 10 μL of sterile water. The PCR amplification conditions were as follows: 2 min at 94 °C, then 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C for PCV2 and 30 s at 72 °C for PCV3, followed by a final elongation step of 10 min at 72 °C. The amplified PCR products for PCV2 and PCV3 were 1057 and 357 bp in length, respectively, as determined by 1% agarose gel electrophoresis.
Amplification of the complete genomic sequences of PCV2 and PCV3
The complete genomic sequences of PCV2 and PCV3 were amplified by PCR using specific primer pairs (Table 1). The whole genomes of 66 PCV2 isolates and four PCV3 isolates were amplified, cloned, and sequenced. Each 50-μL reaction mixture for PCR amplification contained 25 μL of 2× buffer, 10 μL of 2 mM dNTP, 0.5 μL of each primer at a concentration of 10 μM, 2 μL of DNA template, 1 μL of KOD FX Neo polymerase solution (Toyobo, Shanghai, China), and 11 μL of sterile water. The PCR amplification conditions were as follows: 2 min at 94 °C, then 35 cycles of 10 s at 98 °C, 30 s at 55 °C for PCV2 and 30 s at 52 °C for PCV3, and 1 min at 68 °C, followed by a final elongation step of 7 min at 68 °C. The PCR products were analyzed by 1% agarose gel electrophoresis. The PCR products of PCV2 (1766–1768 bp) and PCV3 (2000 bp) were purified using gel extraction kits (Axygen Biotech, Hangzhou, China), according to the manufacturer’s instructions and cloned into the pMD18-T vector (TaKaRa Biotech, Dalian, China). The ligated products were introduced into competent Escherichia coli Top10 cells (Tiangen Biotech, Beijing, China) by transformation. The plasmids were confirmed by restriction enzyme digestion with HindIII for PCV2 and SpeI for PCV3, and then sequenced by Comate Bioscience (Jilin, China) using the primers M13 (-47) and RV-M.
Alignment and phylogenetic analysis
Phylogenetic analysis is regarded as a powerful tool for investigating the evolution of viruses. In this study, multiple alignments were made using the Clustal W of the Molecular Evolutionary Genetics Analysis program, version 7.0 (MEGA7.0), and phylogenetic trees were constructed using the maximum-likelihood (ML) method based on 1000 bootstrap replicates. Alignment analysis was performed with DNAStar (Version 7.0) using the Clustal W method based on the amino acid sequences of ORF1 and ORF2 of PCV2, and ORF2 of PCV3. Details of all sequences used in this study are summarized in Table 2.
Reactivity of mAbs to different PCV2 isolates
In order to test the reactivity of mAbs to the different PCV2 genotypes, nine confirmed positive recombinant plasmids (pMD18-PCV2a-CY29052, PCV2a-YL2801, PCV2a-BX3109, PCV2b-YB2206, PCV2b-HC72302, PCV2b-HD625062, PCV2d-ZB2811, PCV2d-FX3116, and PCV2d-DP2209) were extracted using a TIANprep Midi Plasmid Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. After digestion with HindIII, genomic DNA was separated from plasmid DNA by electrophoresis on a 1% agarose gel. The method for constructing an infectious clone of PCV2 has been reported previously [8]. The rescued PCV2 clone was confirmed using an immunoperoxidase monolayer cell assay (IPMA) as described previously [18]. An immunofluorescence assay (IFA) was used to test the reactivity of different PCV2 mAbs to the rescued PCV2 isolates. Briefly, PK-15 cells in 96-well plates were infected with PCV2a-CY29052, PCV2a-YL2801, PCV2a-BX3109, PCV2b-YB2206, PCV2b-WK01015, PCV2b-HD625062, PCV2d-ZB2811, PCV2d-FX3116, or PCV2d-DP2209 or mock-infected for 48 h, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, and then permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature. Afterward, the cells were incubated at 37°C for 1 h with each of the 13 mAbs (diluted 1:500 in PBS) against PCV2 Cap. After washing three times, the cells were incubated at 37°C for 1 h with goat anti-mouse IgG Abs conjugated with DyLight™ 488 fluorescent dye (diluted 1:500 in PBS; Pierce, Rockford, IL). The fluorescent signals were visualized using an EVOS inverted fluorescence microscope (Life Technologies, Carlsbad, CA, USA). The IFA assays were performed independently three times for each PCV2 isolate.
Restriction fragment length polymorphism (RFLP) analysis of PCV2
In this study, a linearized genome of PCV2 containing a HindIII restriction enzyme site inserted into the viral genome was constructed. Each primer was modified by two nucleotides to create a HindIII restriction enzyme site as a traceable marker (G to A and C to G at positions 996 and 997, respectively). Using this method to linearize the complete PCV2 genomic sequences, the locations of restriction endonuclease sites were identified using Bioedit software (version 7.1.3.0; Biosoft, Cambridge, UK). PCV2a has no AseI site, while PCV2b and PCV2d have only one. The purified PCR-PCV2 products were first digested with AseI to distinguish PCV2a from PCV2b and PCV2d. Based on the analysis of PCV2b and PCV2d, PCV2b has only one SacI site, while PCV2d has one or two. In this way, PCV2d was positively identified when three fragments appeared, but the genotype could not be determined when two fragments appeared after digestion by SacI because PCV2b has only one SacI site. The rescued virus could be differentiated from wild-type PCV2 by PCR and RFLP analysis.
GenBank accession numbers
The complete genomic sequences of the PCV2 and PCV3 isolates analyzed in this study have been submitted to the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) using the Sequin DNA sequence submission tool (https://www.ncbi.nlm.nih.gov/Sequin/).
Results
Prevalence and complete sequences of PCV2 and PCV3
A total of 472 samples collected from diseased and healthy pigs in different geographical areas of China were tested for PCV2 and PCV3 by conventional PCR with specific primers. The prevalence of PCV2, PCV3, and PCV2/3 coinfection was 50.0% (236/472), 13.3% (63/472), and 6.78% (32/472), respectively. The number of PCV2a, b, and d isolates and PCV3 isolates in each province for which complete genome sequences were determined is shown in Fig. 1a and the prevalence of the different PCV2 genotypes by year is shown in Fig. 1b. All of these sequences were submitted to the GenBank database.
a. A total of 66 PCV2 and four PCV3 samples were collected from 10 provinces in China (blue). The colored dots represent the genotypes found in each province. The number of isolates of each genotype is shown with the corresponding colored dot. b. The relationship between the PCV2 subtype and collection year
Phylogenetic analysis based on the complete genome sequences of PCV2
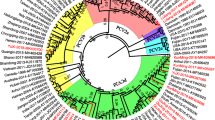
The complete genomic sequences of 66 PCV2 isolates were assigned to three genotypes: PCV2a, PCV2b, and PCV2d. Based on phylogenetic analysis, PCV2a, PCV2b, and PCV2d represented 13.6% (9/66), 25.8% (17/66), and 60.6% (40/66) of the total, respectively (Fig. 2). The genotypes PCV2c, PCV2e, and PCV2f were not found in this study. The within-group sequence identity range for PCV2a, PCV2b, and PCV2d was 96.4%–99.9%, 98.1%–99.9%, and 97.3%–100%, respectively. Moreover, PCV2d, the predominant genotype, shared 94.9%–95.6% and 96.0%–97.3% sequence identity with PCV2a and PCV2b, respectively, while PCV2a and PCV2b shared 95.6%–96.0% sequence identity.
Comparison of PCV2 ORF1 sequences
PCV2 ORF1 encodes the rep and rep’ proteins, which are essential for viral replication and have conserved amino acid sequences. The within-group sequence identity range for PCV2a ORF1, PCV2b ORF1, and PCV2d ORF1 was 99.4%–100%, 98.4%–100%, and 98.1%–100%, respectively. In addition, PCV2d ORF1 shared 98.1%–100% and 98.4%–100% sequence identity with ORF1 of PCV2a and PCV2b, respectively, while PCV2a ORF1 and PCV2b ORF1 shared 98.4%–100% sequence identity. The sixth amino acid of the rep proteins encoded by PCV2a and PCV2b is an asparagine residue, while in PCV2d, it is an asparagine or serine.
Phylogenetic analysis based on PCV2 ORF2
In general, the ML tree based on complete genome sequences of PCV2 isolates was similar to that obtained using ORF2 sequences (Fig. 3). Based on the complete genome sequence, the ORF2 sequence of PCV2-IM1 (intermediate clade, IM1) was close to that of PCV2a and PCV2d [36]. When the phylogenetic tree was constructed based on PCV2 ORF2, the LY2801-IM1 isolate belonged to PCV2d, while it belonged to PCV2a in the tree based on the complete PCV2 sequence. Interestingly, the SD384F16 isolate belonged to PCV2b when the phylogenetic tree was constructed based on PCV2 ORF2, while it belonged to PCV2a in the tree based on the complete sequences and was designated as SD384F16-IM3. The within-group amino acid sequence identity ranges for PCV2a-ORF2, PCV2b-ORF2, and PCV2d-ORF2 were 93.6%–100%, 98.3%–100%, and 97.3%–100%, respectively. In addition, PCV2d-ORF2 shared 88.0%–92.2% and 92.8%–95.2% sequence identity with ORF2 of PCV2a and PCV2b, respectively, while PCV2a-ORF2 and PCV2b-ORF2 shared 91.0%–93.0% sequence identity. The Individual amino acid sequence variations in ORF2 of the different genotypes are listed in Table 3.
Phylogenetic analysis based on the complete genomic sequence of PCV3
Four PCV3 isolates were sequenced and compared to reference isolates of PCV3 and other circoviruses, using MEGA7.0 (Fig. 4). The four PCV3 isolates had a within-group nucleotide sequence identity range of 98.9%–99.7% and 98.8%–99.6% nucleotide sequence identity to isolates five isolates from the USA (PCV3-2164, PCV3-29160, PCV3-US/MO2015, PCV3-US/MN2016, and PCV3-US/SD2016), 99.0%–99.3% nucleotide sequence identity to isolates from the EU, and 98.8%–99.8% nucleotide sequence identity to reference PCV3 isolates from China.
Comparison of PCV3 ORF2 sequences
PCV3 ORF2 contains 645 nucleotides encoding 214 amino acids. Four PCV3 ORF2 sequences shared 98.3%–99.4% nucleotide sequence identity and 98.1%–99.5% amino acid sequence identity. Amino acid 24 and 27 of Cap were crucial for distinguishing genotype PCV3a from PCV3b. When compared to PCV3a, PCV3b contained the amino acid changes A24V and R27K, while PCV3a‐IM contained A24 and K27 [17], indicating that PCV3-CN-JL53/PCV3-CN-LN56, PCV3-CN-HLJ3, and PCV3-CN-0710 belong to genotype PCV3a, PCV3b, and PCV3a-IM, respectively. The four PCV3 Cap sequences identified in this study shared 98.1%–99.5% amino acid and 98.4%–99.4% nucleotide sequence identity, respectively.
Reactivity of mAbs with the rescued PCV2 clones
Nine PCV2 infectious clones were successfully rescued and identified by IPMA (Fig. 5) as described previously [18]. The reactivity of mAbs with the nine PCV2 strains is shown in Table 4. Four of the 13 mAbs (3A5, 8G12, 8E10, and 10D3) reacted with all nine rescued PCV2 isolates, while 1H10, 2C1, 4F5, 4D6, 5B9, 6B10, 5B8, 8E4, and 1D2 reacted with PCV2a-CY29052 and PCV2a-BX3109, but not PCV2-YL2801-IM1, PCV2b-WK01015, PCV2b-YB2206, PCV2d-ZB2811, PCV2d-FX3116, and PCV2d-DP2209. Based on the ORF2 sequence, the reactivity characteristics of PCV2a-YL2801 were predicted to be similar to those of PCV2b and PCV2d isolates.
Nine PCV2 infectious clones were rescued and identified by IPMA. A1, PCV2a-CY29052; A2, PCV2-YL2801-IM1; A3, PCV2a-BX3109; B1, PCV2b-YB2206; B2, PCV2b-WK01015; B3, PCV2b-HD625062; C1, PCV2d-ZB2811; C2, PCV2d-FX3116; C3, PCV2d- DP2209; D, PCV2a-LG (positive control); and E, PK-15 cells (negative control)
RFLP analysis results
An analysis of the PCV2 genome sequence using Bioedit software revealed that the PCV2a isolates had no AseI site, while the PCV2b and PCV2d isolates had only one each. When digested with AseI, PCV2a was represented by one band of 1767–1768 bp, while PCV2b and PCV2d were represented by bands of 511–512 and 1255–1256 bp, respectively (Fig. 6). PCV2a was differentiated from PCV2b/2d by RFLP analysis. PCV2b and PCV2d could be digested by SacI, and PCV2d was distinguished when three fragments of 223, 616, and 928 bp appeared, but not when two fragments of 616 and 1151 bp appeared (Fig. 7). The information of RFLP analysis is summarized in Table 5.
GenBank accession numbers
The complete sequences of PCV2 and PCV3 identified in this study have been submitted to the GenBank database under the accession numbers MK347348 to MK347417 (Table 2).
Discussion
PCV2 is associated with several porcine disease syndromes, collectively known as PCVAD [22]. PCV2 seriously endangers the health of swine and causes tremendous economic losses to the swine industry in China [31, 38]. A high nucleotide substitution rate allows for the continuous evolution of PCV2 and the emergence of novel PCV2 strains. The genotype of PCV2 appears to have shifted twice: PCV2a to PCV2b around 2003 and PCV2b to PCV2d after 2013 [15, 39]. Recently, PCV2d was recognized as the predominant genotype worldwide. Although vaccines based on PCV2a could provide protection against infection with PCV2b and PCV2d [23], substantial genetic divergence among PCV2d strains over time, which might have changed the pathogenicity of the virus, has been observed in PCV2 isolates collected from farms where the vaccine program had failed.
Since the first report of PCV3 in the USA in 2016, many studies have explored the epidemiology and source of the virus. Recently, PCV3 was successfully rescued and propagated in PK-15 cells for 15 passages, and this virus was capable of reproducing severe PDNS-like disease [14]. A high prevalence of PCV3 (44.2% [159/360]) has been reported in South Korea [15]. In northeast China, the total PCV3-positive rate was 33.3% (35/105), with rates of 17.8% (8/45), 66.7% (10/15), and 37.8% (17/45) in Hei Longjiang, Ji Lin, and Liao Ning provinces, respectively [12]. However, Qi et al. reported that the PCV3 positive rate in China was 12.2% (75/616) at the sample level and 24.1% (42/174) at the farm level [27]. In the present study, the PCV3-positive rate was lower 13.3% (63/472) at the sample level than in earlier studies in China, but is consistent with the results reported by Qi et al. in 2018 [27]. Moreover, Zhai et al. reported that the detection rate of PCV3 in the respiratory tract was much higher than that in the digestive tract [39]. The detection rate of PCV3 is thus closely related to the tissue of origin. A pair of PCV3-specific primers was used for PCR amplification of the complete PCV3 genome sequence, and a one-step PCR technique was established in our laboratory for amplification of PCV3. In the present study, we referred to the methods described by Li et al. to divide the PCV3 strains into different clades [17]. Based on the amino acid sequence of PCV3 Cap, four complete PCV3 sequences were obtained: PCV3-CN-JL53/PCV3-CN-LN56, PCV3-CN-HLJ3, and PCV3-CN-0710, which belonged to PCV3a, PCV3b, and PCV3a-IM, respectively. However, at present, there are no universally recognized rules for the classification of PCV3, as more complete sequences are needed to establish the criteria to be used. In this study, 50.0% (236/472) of the samples collected from 2015 to 2018 were positive for PCV2. In a previous retrospective study conducted in China, using samples collected from 1999 to 2017, the PCV2-positive rate was 54.31% (227/418) [37]. Qu et al. reported that, out of 674 samples collected from 2006 to 2016 in Hunan province, 62% were positive for that PCV2 and PCV2d had become the predominant genotype beginning in 2008 [28]. In this study, the complete genome sequences of 66 PCV2 isolates were determined, with PCV2a, PCV2b, and PCV2d representing 13.6% (9/66), 25.8% (17/66), and 60.6% (40/66) of the total respectively. The three main genotypes of PCV2 were widespread on pig farms, while PCV2d was the predominant PCV2 genotype in China. PCV2d has been reported to be the predominant genotype by several research groups [15, 28, 38].
The use of mAbs to identify the antigenic subtype of PCV2 has been reported previously [29, 30]. In this study, nine PCV2 isolates was rescued by the reverse genetics method described by Liu et al. [18]. Because thirteen mAbs were generated against PCV2a Cap, they reacted more strongly to the PCV2a subtype than to PCV2b and PCV2d. The mAbs 2C1, 3A5, 4F5, 4D6, 5B8, and 8E4 reacted with 2a-CY29052, BX3109, and PCV2a-LG (positive control). Based on phylogenetic analysis of complete sequences, the PCV2-YL2801-IM1 isolate belonged to the PCV2a genotype. However, the sequence of ORF2 was most similar to that of PCV2d. Based on this characteristic, the reaction characteristics of PCV2-YL2801-IM1 with thirteen mAbs were similar to those of PCV2b and PCV2d. In comparison to the amino acid sequences of PCV2a with PCV2b and PCV2d, the mutations A(59)R/K, T(86)S, K(88)P, I(91)V, P(151)T, S(190)A/T, K/A/R(191)G, and K/T(206)I may be the key amino acids for the change of the reaction characteristics of mAbs.
PCR-RFLP is a relatively simple and rapid method that is expensive than sequencing for genotyping, allowingPCV2 isolates from a large number of pigs on farms to be be analyzed regularly. A genotypic change to PCV2 was revealed by RFLP analysis based on PCV2 ORF2, as reported previously by Takahagi et al. [36]. In this study, genotyping of PCV2 isolates based on RFLP analysis was performed using complete PCV2 sequences. We developed an RFLP method requiring only two restriction endonucleases (AseI and SacI). First, AseI was employed to discriminate PCV2a from PCV2b and PCV2d. Then, SacI was used to discriminate PCV2b from PCV2d, based on whether three fragments were produced, as mentioned above. When two fragments were observed, the use of SacI was insufficient to discriminate PCV2b from PCV2d. The PCV2 genome should be further analyzed to identify more suitable restriction sites to distinguish different PCV2 genotypes.
In view of the great harm that PCV2 and PCV3 cause to the pig industry, it is necessary to monitor changes in the viral genome sequence and explore the impact of such changes on viral virulence. In order to investigate the prevalence of PCV2 and PCV3 in China from 2015 to 2018, a total of 472 samples collected from different regions of China were tested for PCV2 and PCV3. The complete genome sequences of 66 PCV2 isolates were determined, with PCV2a, PCV2b, and PCV2d representing 13.6% (9/66), 25.8% (17/66), and 60.6% (40/66) of the total, respectively. The three main genotypes of PCV2 were widespread on pig farms, while PCV2d was the predominant PCV2 genotype in China. The epidemic trend and pathogenicity of PCV3 should be continuously monitored. Overall, the results of the present study provide detailed information regarding the prevalence, genetic diversity, and molecular epidemiology of PCV2 and PCV3, which should prove useful for future efforts to develop effective preventive and control measures.
References
Bao F, Mi S, Luo Q, Guo H, Tu C, Zhu G, Wang W (2017) Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound Emerg Dis 65(2):432–440
Chen GH, Mai KJ, Zhou L, Wu RT, Tang XY, Wu JL, He LL, Lan T, Xie QM, Sun Y, Ma JY (2017) Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg Dis 64(6):1650–1654
Cheung AK (2015) Specific functions of the Rep and Rep proteins of porcine circovirus during copy-release and rolling-circle DNA replication. Virology 481:43–50
Collins PJ, McKillen J, Allan G (2017) Porcine circovirus type 3 in the UK. Vet Rec 181:599
Davies B, Wang X, Dvorak CMT, Marthaler D, Murtaugh MP (2016) Diagnostic phylogenetics reveals a new Porcine circovirus 2 cluster. Virus Res 217:32–37
Faccini S, Barbieri I, Gilioli A, Sala G, Gibelli LR, Moreno A, Sacchi C, Rosignoli C, Franzini G, Nigrelli A (2017) Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound Emerg Dis 64(6):1661–1664
Franzo G, Segalés J (2018) Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS One 13(12):e0208585
Guo LJ, Lu YH, Huang LP, Wei YW, Wu HL, Liu CM (2011) First construction of infectious clone for newly emerging mutation porcine circovirus type 2 (PCV2) followed by comparison with PCV2a and PCV2b genotypes in biological characteristics in vitro. Virol J 8:291
Guo LJ, Lu YH, Wei YW, Huang LP, Liu CM (2010) Porcine circovirus type 2 (PCV2): genetic variation and newly emerging genotypes in China. Virol J 7:273
Hayashi S, Ohshima Y, Furuya Y, Nagao A, Oroku K, Tsutsumi N, Sasakawa C, Sato T (2018) First detection of porcine circovirus type 3 in Japan. J Vet Med Sci 80(9):1468–1472
Harmon KM, Gauger PC, Zhang J, Piñeyro PE, Dunn DD, Chriswell AJ (2015) Whole-genome sequences of novel porcine circovirus type 2 viruses detected in swine from Mexico and the United States. Genome Announc 3(6):e01315
Ha Z, Xie CZ, Li JF, Wen SB, Zhang KL, Nan FL, Zhang H, Guo YC, Wang W, Lu HJ, Jin NY (2018) Molecular detection and genomic characterization of porcine circovirus 3 in pigs from Northeast China. BMC Vet Res 14(1):321
He J, Cao J, Zhou N, Jin Y, Wu J, Zhou J (2013) Identification and functional analysis of the novel ORF4 protein encoded by porcine circovirus type 2. J Virol 87(3):1420–1429
Jiang H, Wang D, Wang J, Zhu S, She R, Ren X, Tian J, Quan R, Hou L, Li Z, Chu J, Guo Y, Xi Y, Song H, Yuan F, Wei L, Liu J (2018) Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol 93(4):e02045-18
Kwon T, Yoo SJ, Park CK, Lyoo YS (2017) Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol 207:178–180
Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q (2017) Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis 64(3):703–708
Li G, He W, Zhu H, Bi Y, Wang R, Xing G, Zhang C, Zhou J, Yuen KY, Gao GF, Su S (2018) Origin, genetic diversity, and evolutionary dynamics of novel porcine Circovirus 3. Adv Sci 5(9):1800275
Liu C, Wei Y, Zhang C, Lu Y, Kong X (2007) Construction and characterization of porcine circovirus type 2 carrying a genetic marker strain. Virus Res 127(1):95–99
Liu J, Chen I, Du Q, Chua H, Kwang J (2006) The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J Virol 80:5065–5073
Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM (1998) Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol 79(9):2171–2179
Nawagitgul P, Morozov I, Bolin SR, Harms FA, Sorden SD, Paul PS (2000) Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J Gen Virol 81(9):2281–2287
Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM (2007) Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest 19(6):591–615
Opriessnig T, Xiao CT, Halbur PG, Gerber PF, Matzinger SR, Meng XJ (2017) A commercial porcine circovirus (PCV) type 2a-based vaccine reduces PCV2d viremia and shedding and prevents PCV2d transmission to naive pigs under experimental conditions. Vaccine 35(2):248–254
Palinski R, Pineyro P, Shang P, Yuan F, GuoR Fang Y, Byers E, Hause BM (2016) A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol 91(1):01879-16
Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E (2016) Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J 13(1):184
Pringle CR (1999) Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia. Adv Virol 144:2065–2070
Qi S, Su M, Guo D, Li C, Wei S, Feng L, Sun D (2019) Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 Provinces of China during 2015–2017. Transbound Emerg Dis 66(2):1004–1015
Qu T, Li R, Yan M, Luo B, Yang T, Yu X (2018) High prevalence of PCV2d in Hunan province, China: a retrospective analysis of samples collected from 2006 to 2016. Adv Virol 163(7):1897–1906
Saha D, Huang L, Bussalleu E, Lefebvre DJ, Fort M, Van Doorsselaere J, Nauwynck HJ (2012) Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Vet Microbiol 157(1–2):13–22
Shang SB, Jin YL, Jiang XT, Zhou JY, Zhang X, Xing G, He JL, Yan Y (2009) Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol Immunol 46(3):327–334
Shuai J, Wei W, Li X, Chen N, Zhang Z, Chen X, Fang W (2007) Genetic characterization of porcine circovirus type 2 (pcv2) from pigs in high-seroprevalence areas in southeastern China. Virus Genes 35(3):619–627
Segalés J, Olvera A, Grau-Roma L, Charreyre C, Nauwynck H, Larsen L, Dupont K, McCullough K, Ellis J, Krakowka S, Mankertz A, Fredholm M, Fossum C, Timmusk S, Stockhofe-Zurwieden N, Beattie V, Armstrong D, Grassland B, Baekbo P, Allan G (2008) PCV-2 genotype definition and nomenclature. Vet Rec 162(26):867–868
Stadejek T, Wozniak A, Milek D, Biernacka K (2017) First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis 64(5):1350–1353
Tischer I, Rasch R, Tochtermann G (1974) Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A 226(2):153–156
Tochetto C, Lima DA, Varela APM, Loiko MR, Paim WP, Scheffer CM, Herpich JI, Cerva C, Schmitd C, Cibulski SP, Santos AC, Mayer FQ, Roehe PM (2018) Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound Emerg Dis 65(1):5–9
Takahagi Y, Nishiyama Y, Toki S, Yonekita T, Morimatsu F, Murakami H (2008) Genotypic change of porcine circovirus type 2 on japanese pig farms as revealed by restriction fragment length polymorphism analysis. J Vet Med Sci 70(6):603–606
Wang H, Gu J, Xing G, Qiu X, An S, Wang Y, Zhang C, Liu C, Gong W, Tu C, Su S, Zhou J (2018) Genetic diversity of porcine circovirus type 2 in China between 1999–2017. Transbound Emerg Dis 66(1):599–605
Xiao CT, Halbur PG, Opriessnig T (2015) Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J Gen Virol 96(7):1830–1841
Zhai SL, Chen SN, Xu ZH, Tang MH, Wang FG, Li XJ, Sun BB, Deng SF, Hu J, Lv DH, Wen XH, Yuan J, Luo ML, Wei WK (2014) Porcine circovirus type 2 in China: an update on and insights to its prevalence and control. Virol J 11:88
Zhang L, Luo Y, Liang L, Li J, Cui S (2018) Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in china detected by duplex nanoparticle-assisted pcr. Infect Genet Evol 60:1–6
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant no. 31873012) and the National Key Research and Development Programs (Grant nos. 2017YFD0500903 and 2017YFD0501603)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Sheela Ramamoorthy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xia, D., Huang, L., Xie, Y. et al. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch Virol 164, 2435–2449 (2019). https://doi.org/10.1007/s00705-019-04336-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04336-4