Abstract
Two begomoviruses were isolated in the northern Brazilian state of Pará, infecting non-cultivated Hibiscus sp. and cultivated tomato (Solanum lycopersicum). The complete genomes (DNA-A and DNA-B) of the two viruses showed the typical organization of New World bipartite begomoviruses. Based on the species assignment criteria in the genus Begomovirus, each virus is a member of a new species. The virus from Hibiscus is most closely related to sida yellow mosaic Yucatan virus, while the tomato virus is most closely related to abutilon mosaic Brazil virus and corchorus mottle virus. Recombination events were detected in the DNA-A of the tomato virus, but not in the Hibiscus virus genome. We propose the names “hibiscus golden mosaic virus” (HGMV) and “tomato chlorotic leaf curl virus” (ToCLCV) for the viruses reported in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus Begomovirus (family Geminiviridae) includes viruses that infect dicotyledonous plants and whose genomes are composed of one or two genomic components of circular, single-strand DNA (ssDNA) of about 2,700 nucleotides, encapsidated in icosahedral particles and transmitted by whiteflies of the Bemisia tabaci cryptic species complex [33]. Begomoviruses constitute an important group of plant pathogens responsible for severe diseases in several crops of economic importance worldwide [27]. The two genomic components of bipartite viruses are designated as DNA-A and DNA-B. The DNA-A contains genes for replication and encapsidation of genetic material. The DNA-B is responsible for intra- and intercellular movement in plant. Both components are required for systemic infection [26].
Brazil is a center of diversity of begomoviruses, with a great number of these viruses reported in non-cultivated hosts [4, 5, 9] as well as in crops [12, 25]. The most important diseases are bean golden mosaic caused by bean golden mosaic virus (BGMV) and golden mosaic/leaf curl diseases caused by a complex of begomoviruses in tomato [27]. The emergence of tomato-infecting begomoviruses in Brazil has been explained by horizontal transfer of indigenous viruses infecting non-cultivated hosts following the introduction of polyphagous Bemisia tabaci Middle East Asia Minor 1 (MEAM-1) in the early 1990s [24, 25]. A large diversity of begomoviruses has been sampled in non-cultivated hosts [2, 4, 9, 10, 13, 15, 17, 20,21,22,23, 29, 30].
The majority of begomoviruses from Brazil were isolate in the central, northeastern, and southeastern regions of the country [12]. These areas have a climate that favors the buildup of whitefly populations, with a well-defined dry winter season, which is also relatively warm. The northern region, which includes the Amazon forest, comprises almost 50% of the country’s area and has an equatorial climate without a dry season. Possibly as a result, whitefly populations rarely reach epidemic levels, and begomoviruses have not been reported. Nevertheless, recent years have seen atypical dry spells in the Amazon, which have been attributed to climate change [8, 32].
In June 2016, two tomato (Solanum lycopersicum) samples with mosaic and leaf distortion (Fig. 1A) were collected in a field near the city of Altamira (S3°16′094′′, W52°23′38.4′′), and in August of the same year, one Hibiscus sp. plant with yellow mosaic symptoms (Fig. 1B) was collected in the city of Igarapé Miri (S1°58′30′′, W48°57′35′′), both in the state of Pará, northern Brazil. The identity of the Hibiscus sample was confirmed by DNA barcoding using the chloroplast matK and rbcL genes. Total DNA was extracted from each sample [6] and used as a template for rolling-circle amplification (RCA) using the phi29 DNA polymerase [11]. Amplification products were individually cleaved with SacI (tomato samples) and EcoRI/HindIII (Hibiscus sample), ligated into the pBLUESCRIPT-KS+ (pKS+) plasmid vector (Stratagene) previously cleaved with the same enzyme, and introduced into Escherichia coli DH5α by transformation. Viral inserts were sequenced commercially (Macrogen, Inc.). Full-length genomes were assembled using Geneious v. 8.1 [14]. Sequences were initially analyzed using the BLASTn algorithm [1], and sequence identity to the closest begomoviruses was calculated with Species Demarcation Tool v.1.2 (SDT) [18]. Full-length genomes of begomoviruses were aligned with MUSCLE [7] implemented in MEGA v. 7.0 [31]. Phylogenetic trees based on DNA-A and DNA-B alignments were generated by Bayesian inference using MrBayes v. 3.2.6 [28] with the GTR + G+I nucleotide substitution model selected by MrModeltest v. 2.2 [19] in the Akaike information criterion (AIC). Analyses were carried out for 20,000,000 generations, and sampling was done at every 1,000 steps to produce the distribution tree. The first 4,000 trees were discarded. The trees were visualized in FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree) and edited in CorelDRAW 2017. To detect putative recombination events, the software RDP4 [16] was used. Recombination events were considered reliable only if they were detected by at least four of the seven methods implemented in the program.
DNA-A and DNA-B components were cloned from the Hibiscus sp. and tomato samples. All components have the conserved nonanucleotide (5′-TAATATTAC-3′) located at the origin of replication. DNA components from the same sample have identical iteron sequences (GGTGT for the components from the tomato samples; GGTGG for the components from the Hibiscus sp. sample), and digestion of the RCA products from each sample with a 4-base cutter restriction enzyme generated fragments with lengths adding up to approximately 5,200 nt (data not shown). These results indicate that they were cognate DNA components, and they were the only two DNA components present in each sample. All components have the typical organization of New World, bipartite begomoviruses, with five ORFs in the DNA-A and two ORFs in the DNA-B.
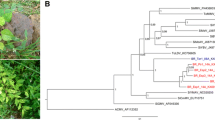
Three clones (two DNA-A and one DNA-B) were sequenced from the tomato samples. The DNA-A clones corresponding to isolates BR-Alt1-16 (2612 nt; GenBank accession number MK558058) and BR-Alt3-16 (2615 nt; MK558060) (from different samples) showed 99.62% sequence identity amongst themselves and a maximum sequence identity of 84% with the DNA-A of abutilon mosaic Brazil virus (AbMBV; JF694482) and corchorus mottle virus (CoMoV; JQ805781) (Fig. 2A). Based on the begomovirus species demarcation criterion established by the International Committee on Taxonomy of Viruses (ICTV), which considers a 91% identity cutoff for the full-length DNA-A component [3], these isolates should be considered members of a new species, for which we propose the name “Tomato chlorotic leaf curl virus” (ToCLCV). In the DNA-A phylogenetic tree, ToCLCV grouped in a clade with viruses from Central and North America (Fig. 2B), which also includes viruses from Brazil that have been shown to be members of a lineage of New World begomoviruses that is distinct from other Brazilian begomoviruses [25]. The DNA-B tree shows the same topology (data not shown). Recombination analysis indicated a recombination event in the DNA-A of BR-Alt1-16 and BR-Alt3-16, with putative recombination breakpoints at positions 204 and 969, with tomato leaf distortion virus (KC706605) and sida chlorotic vein virus (KX691405) inferred as the major and minor parent, respectively.
A. Nucleotide identity matrix (DNA-A) of isolates BR-IgM1-16 (hibiscus golden mosaic virus), BR-Alt1-16 and BR-Alt3-16 (tomato chlorotic leaf curl virus) with the most closely related begomoviruses. Identities were calculated with SDT v. 1.2. B. Phylogenetic reconstruction using Bayesian inference. The midpoint-rooted tree is based on the complete DNA-A sequences of the begomoviruses described in this work and other New World begomoviruses. Numbers at the nodes indicate Bayesian posterior probabilities
Pairwise sequence comparisons with the DNA-A of isolate BR-IgM1-16 (MK558061) from Hibiscus sp. indicated a maximum nucleotide sequence identity of 79% to sida yellow mosaic Yucatan virus (SiYMYuV; DQ875872) (Fig. 2A). Thus, the isolate BR-IgM1-16 should be considered a member of a new species, whose proposed name is “Hibiscus golden mosaic virus” (HGMV). A phylogenetic tree based on DNA-A nucleotide sequences indicates a distant relationship of HGMV to SiYMYuV and other begomoviruses from Central/North America and the Caribbean (Fig. 2B). The HGMV DNA-B (MK558062) is most closely related to those of blainvillea yellow spot virus and tomato common mosaic virus (data not shown). Recombination analysis revealed no evidence of significant recombination events.
Weeds and wild plants can serve as reservoirs of viruses for cultivated plants, especially between growing seasons. In addition, the great biodiversity of begomoviruses in non-cultivated plants may contribute to the emergence of new viruses in cultivated plants. Therefore, the characterization of begomoviruses in non-cultivated hosts provides information about the possible threat of new epidemics.
Intensive sampling over the last 20 years has led to the description of several new begomoviruses infecting cultivated (especially tomato) and non-cultivated plants in Brazil. However, the Brazilian northern region has been underrepresented in these surveys, and the true extent of begomovirus diversity in this region is likely to have been underestimated as well. The two new viruses described here are more closely related to begomoviruses from Central/North America and the Caribbean than to begomoviruses from South America. Additional sampling in this geographically large region is necessary to illuminate the true richness of begomoviruses present in its different biomes, as well as their evolutionary relationships.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Blawid R, Fontenele RS, Lacorte C, Ribeiro SG (2013) Molecular and biological characterization of corchorus mottle virus, a new begomovirus from Brazil. Arch Virol 158:2603–2609
Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JC, Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A (2015) Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160:1593–1619
Castillo-Urquiza GP, Beserra JEA Jr, Bruckner FP, Lima ATM, Varsani A, Alfenas-Zerbini P, Zerbini FM (2008) Six novel begomoviruses infecting tomato and associated weeds in Southeastern Brazil. Arch Virol 153:1985–1989
Costa AS, Carvalho AMB (1960) Comparative studies between Abutilon and Euphorbia mosaic viruses. J Phytopathol 38:129–152
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull 19:11–15
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:1–19
Erfanian A, Wang G, Fomenko L (2017) Unprecedented drought over tropical South America in 2016: significantly under-predicted by tropical SST. Sci Rep 7:5811
Ferro CG, Silva JP, Xavier CAD, Godinho MT, Lima ATM, Mar TB, Lau D, Zerbini FM (2017) The ever increasing diversity of begomoviruses infecting non-cultivated hosts: new species from Sida spp. and Leonurus sibiricus, plus two New World alphasatellites. Ann Appl Biol 170:204–218
Fiallo-Olivé E, Zerbini FM, Navas-Castillo J (2015) Complete nucleotide sequences of two new begomoviruses infecting the wild malvaceous plant Melochia sp. in Brazil. Arch Virol 160:3161–3164
Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T (2004) A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J Virol Methods 116:209–211
Inoue-Nagata AK, Lima MF, Gilbertson RL (2016) A review of geminivirus diseases in vegetables and other crops in Brazil: current status and approaches for management. Hortic Bras 34:8–18
Jovel J, Reski G, Rothenstein D, Ringel M, Frischmuth T, Jeske H (2004) Sida micrantha mosaic is associated with a complex infection of begomoviruses different from Abutilon mosaic virus. Arch Virol 149:829–841
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Mar TB, Xavier CAD, Lima ATM, Nogueira AM, Silva JCF, Ramos-Sobrinho R, Lau D, Zerbini FM (2017) Genetic variability and population structure of the New World begomovirus Euphorbia yellow mosaic virus. J Gen Virol 98:1537–1551
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Melo AM, Silva SJ, Ramos-Sobrinho R, Ferro MM, Assuncao IP, Lima GS (2016) Cnidoscolus mosaic leaf deformation virus: a novel begomovirus infecting euphorbiaceous plants in Brazil. Arch Virol 161:2605–2608
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277
Nylander JAA (2004) MrModeltest v2. Program distributed by the author Evolutionary Biology Centre. Uppsala University, Uppsala
Paprotka T, Metzler V, Jeske H (2010) The complete nucleotide sequence of a new bipartite begomovirus from Brazil infecting Abutilon. Arch Virol 155:813–816
Passos LS, Rodrigues JS, Soares ÉCS, Silva JP, Zerbini FM, Araújo ASF, Beserra JEA (2017) Complete genome sequence of a new bipartite begomovirus infecting Macroptilium lathyroides in Brazil. Arch Virol 162:3551–3554
Passos LS, Teixeira JWM, Teixeira KJML, Xavier CAD, Zerbini FM, Araújo ASF, Beserra JEA (2017) Two new begomoviruses that infect non-cultivated malvaceae in Brazil. Arch Virol 162:1795–1797
Pinto VB, Silva JP, Fiallo-Olivé E, Navas-Castillo J, Zerbini FM (2016) Novel begomoviruses recovered from Pavonia sp. in Brazil. Arch Virol 161:735–739
Ribeiro SG, Ávila AC, Bezerra IC, Fernandes JJ, Faria JC, Lima MF, Gilbertson RL, Zambolim EM, Zerbini FM (1998) Widespread occurrence of tomato geminiviruses in Brazil, associated with the new biotype of the whitefly vector. Plant Dis 82:830
Rocha CS, Castillo-Urquiza GP, Lima ATM, Silva FN, Xavier CAD, Hora-Junior BT, Beserra-Junior JEA, Malta AWO, Martin DP, Varsani A, Alfenas-Zerbini P, Mizubuti ESG, Zerbini FM (2013) Brazilian begomovirus populations are highly recombinant, rapidly evolving, and segregated based on geographical location. J Virol 87:5784–5799
Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394
Rojas MR, Macedo MA, Maliano MR, Soto-Aguilar M, Souza JO, Gilbertson RL, Briddon RW, Kenyon LA, Bustamante RFR, Zerbini FM, Adkins S, Legg JP, Kvarnheden A, Wintermantel WM, Sudarshana MR, Peterschmitt M, Lapidot M, Martin DP, Moriones E, Inoue-Nagata AK (2018) World management of geminiviruses. Annu Rev Phytopathol 56:637–677
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Silva SJC, Castillo-Urquiza GP, Hora-Júnior BT, Assunção IP, Lima GSA, Pio-Ribeiro G, Mizubuti ESG, Zerbini FM (2011) High genetic variability and recombination in a begomovirus population infecting the ubiquitous weed Cleome affinis in northeastern Brazil. Arch Virol 156:2205–2213
Silva SJC, Castillo-Urquiza GP, Hora-Junior BT, Assunção IP, Lima GSA, Pio-Ribeiro G, Mizubuti ESG, Zerbini FM (2012) Species diversity, phylogeny and genetic variability of begomovirus populations infecting leguminous weeds in northeastern Brazil. Plant Pathol 61:457–467
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Yang J, Tian H, Pan S, Chen G, Zhang B, Dangal S (2018) Amazon drought and forest response: largely reduced forest photosynthesis but slightly increased canopy greenness during the extreme drought of 2015/2016. Global Change Biol 24:1919–1934
Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Varsani A, ICTV Consortium (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131–133
Funding
This work was funded by Norte Energia S.A., CAPES (Finance Code 001), CNPq (Grant 409599/2016-6 to FMZ) and FAPEMIG (Grant APQ-03444-16 to FMZ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Jesús Navas-Castillo.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The sequences described in this work have been deposited in GenBank, with accession numbers MK558058–MK558062.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Quadros, A.F.F., Silva, J.P., Xavier, C.A.D. et al. Two new begomoviruses infecting tomato and Hibiscus sp. in the Amazon region of Brazil. Arch Virol 164, 1897–1901 (2019). https://doi.org/10.1007/s00705-019-04245-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04245-6