Abstract
Here, we report a novel member of the genus Polerovirus, zucchini aphid-borne yellows virus (ZABYV), which was identified in zucchini grown for seed production in the Xinjiang Uygur Autonomous Region, China. The complete nucleotide sequence of the ZABYV genome was determined and found to be 5,792 nucleotides in length, and like those of other poleroviruses, to contain seven open reading frames (ORFs). Multiple sequence alignments and phylogenetic analysis indicated that ZABYV is a new member of the genus Polerovirus, although several regions of its genome are closely related to chickpea chlorotic stunt virus (CpCSV). Further comparative analysis suggested that ZABYV originated from a recombination event between CpCSV and another unknown virus in the genus Polerovirus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Zucchini (Cucurbita pepo) is an annual herbaceous crop that belongs to the genus Cucurbita of family Cucurbitaceae. Its young fruit is favored by consumers all over the world as a fresh vegetable. Mature seeds of cultivars with large-grain-type kernels are processed as food or food raw materials. Most of the zucchini consumed as seeds are planted in the Xinjiang Uygur Autonomous Region in China as one of its economically important crops. Viral diseases of zucchini in this region are one of the major factors limiting its production. There are two types of viral diseases on Cucurbitaceae crops in the Xinjiang Uygur Autonomous Region: mosaic virus disease, caused by zucchini yellow mosaic virus (ZYMV), watermelon mosaic virus (WMV) and papaya ring spot virus (PRSV) of the genus Potyvirus and cucumber mosaic virus (CMV) of the genus Cucumovirus, and yellowing virus disease, caused by cucurbit aphid-borne yellows virus (CABYV) and melon aphid-borne yellows virus (MABYV) of the genus Polerovirus and cucurbit chlorotic yellows virus (CCYV) of the genus Crinivirus [8, 9, 11].
According to the ICTV Master Species List (https://talk.ictvonline.org/files/master-species-lists/), the genus Polerovirus of the family Luteoviridae, includes 19 virus species. Particles of these viruses are icosahedral, and their genomes are positive-sense, single-stranded RNAs that have a genome-linked protein (VPg) linked to their 5’-terminal ends. The genome contains seven open reading frames (ORFs). ORF0 encodes a protein with a silencing suppressor function, ORF1 encodes the VPg and protease, part of ORF1 and ORF2 encode an RNA-dependent RNA polymerase (RDRP) via a -1 ribosomal frameshift, ORF3 encodes the major coat protein (CP), ORF3a starts with a non-AUG start codon and encodes a protein involved in long-distance movement of the virus, ORF4 encodes a movement protein (MP), and ORF3 and ORF5, via a readthrough of a termination codon at end of ORF3, encodes a minor coat protein and a possible aphid transmission and virion stability factor [2]. Six members of in the genus Polerovirus, including CABYV, MABYV, suakwa aphid-borne yellows virus (SABYV) [10], pepo aphid-borne yellows virus (PABYV) [1], cucumber aphid-borne yellows virus (CuABYV) and luffa aphid-borne yellows virus (LABYV) [4], have been reported to infect crops of the family Cucurbitaceae. These viruses are transmitted in a persistent, circulative, and non-propagative manner by aphids, inducing yellowing and chlorosis of leaves in Cucurbitaceae crops.
In August 2016, zucchini grown for seed production in Fukang County, Wujiaqu City, Xinjiang Uygur Autonomous Region, was severely affected by viral diseases, with an average incidence rate of 80%. Most of the diseased plants showed a mixture of mosaic and yellowing symptoms (Fig. 1A). We collected six virus-infected samples from two zucchini plots in this region. Of these six samples, one and five were identified using DAS-ELISA (Agdia, Elkhart, USA) to be infected by ZYMV and WMV, respectively, whereas none were infected by PRSV or CMV (Supplementary Table 1). Total RNA was extracted from these six samples using an RNAprep Pure Plant Kit (Tiangen, Beijing, China). Three samples (XJ2, XJ3 and XJ5) were identified to be polerovirus positive (Supplementary Table 1) using a One Step RT-PCR Kit (Takara, Dalian, China) with the polerovirus universal primer pair Pol-G-F and Pol-G-R [3] (Supplementary Table 2), whereas the expected PCR products were not obtained using species-specific primer pairs for CABYV (CA3414F and Pococp R) [12], MABYV (MA3669F and Pococp R) [12] or CCYV (CCYV-CPF and CCYV-CPR) [8]. PCR products from the three polerovirus-positive samples were cloned into the pTOPO vector using a CV15-Zero Background pTOPO-TA Simple Cloning Kit (Aidlab, Beijing, China), and their sequences were determined using Sanger dideoxy DNA sequencing (Sangon, Shanghai, China). The nucleotide sequences of these PCR products from the three samples were 99.8% identical. Comparison of these sequences against the GenBank nucleotide (nt) database revealed that they shared 70-75% nucleotide sequence identity with members of the genus Polerovirus, with the highest similarity to turnip yellows virus (TuYV; GenBank accession no. KU198395), indicating that this virus might be a novel member of the genus Polerovirus. We named this virus “zucchini aphid-borne yellows virus” (ZABYV).
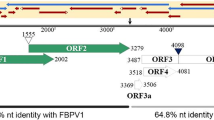
Characterization of zucchini aphid-borne yellows virus (ZABYV), a novel member of the genus Polerovirus, isolated from zucchini grown for seed production in China. (A) Symptoms on the leaf of the zucchini plant from which ZABYV was isolated. (B) Genome organization of ZABYV and the complete sequencing and cloning strategy used in this study. Rectangles represent predicted ORFs, whose positions in the genome are shown. A triangle, a cross, and a star represent the -1 frameshift site, the non-AUG start codon, and the stop codon that is read through in the ZABYV genome, respectively. Arrows represent the PCR fragments that were cloned into pTOPO, together covering the entire genome of ZABYV. The primers used for amplifying the PCR fragments are indicated at the ends of each arrow. The region between the dotted lines is the recombinant region of the ZABYV genome. (C) Phylogenetic trees of ZABYV and other members in the genus Polerovirus based on the complete genome sequence (left) and the amino acid sequences of RDRP (middle) and CP (right). Barley yellow dwarf virus (BYDV, genus Luteovirus) and pea enation mosaic virus (PEMV, genus Enamovirus) were used as outgroups. The following members of the genus Polerovirus were included:chickpea chlorotic stunt virus (CpCSV), white clove mottle virus (WhCMV), suakwa aphid-borne yellows virus (SABYV), pepo aphid-borne yellows virus (PABYV), cucurbit aphid-borne yellows virus (CABYV), melon aphid-borne yellows virus (MABYV), cotton leafroll dwarf virus (CLRDV), luffa aphid-borne yellows virus (LABYV), pepper vein yellows virus (PeVYV), tobacco vein distorting virus (TVDV), cereal yellow dwarf virus (CYDV), potato leafroll virus (PLRV), carrot red leaf virus (CtRLV), brassica yellows virus (BrYV), turnip yellows virus (TuYV), beet chlorosis virus (BChV), beet mild yellowing virus (BMYV), beet western yellows virus (BWYV), maize yellow dwarf virus (MYDV), and sugarcane yellow leaf virus (ScYLV). GenBank accession numbers are shown on the tree after the virus names. Bootstrap values less than 70 are not shown. (D) Recombination in the ZABYV genome. The pink region, which shows opposite sequence identity patterns between ZABYV and BrYV, and between ZABYV and CpCSV, compared to other regions in the ZABYV genome, represents the recombinant region in the ZABYV genome
Total RNA of sample XJ5 was subjected to rRNA-depletion treatment using a Ribo-ZeroTM Magnetic Kit (Illumina) according to the manufacturer’s instructions. An RNA-Seq library was then constructed using a Next Ultra Directional RNA Library Prep Kit (NEB, Ipswich, USA) and sequenced on an Illumina HiSeq X10 platform. After removing the adapter sequence and low-quality reads from the raw sequencing data, we obtained a total of 68.5 million high-quality cleaned reads. We then applied VirusDetect (Version 1.7) [13] to the cleaned RNA-Seq reads for virus identification. The plant virus database (provided within VirusDetect) was used as a reference for reference-guided assembly, and the zucchini genome sequence [7] was used to subtract RNA-Seq reads derived from the host. Among the final assembled contigs, in addition to a long contig that could be aligned to the WMV genome (GenBank accession no. EU660585) with 99% coverage and 99.3% identity, we also obtained eight contigs (ranging from 213 nt to 1483 nt in length) that could be aligned to genomes of CpCSV (GenBank accession no. JF507725), TuYV (GenBank accession nos. KR706247, X13063, ALL26142 and CAA31463) and cotton leafroll dwarf virus (CLRDV; GenBank accession nos. AKU41568) by blastn or blastx (Supplementary Table 3).
To obtain the complete genome sequence of ZABYV, 10 primers were designed based on the sequence information of the eight contigs. Four of them were nested primers (5R477 and 5R744; 3R5171 and 3R5260) for 5’ and 3’ RACE (rapid amplification of cDNA ends), and the other three primer pairs (P458F and P2830R; P2813F and P5278R; P4672F and P5646R) were used to amplify the middle part of the genome (Supplementary Table 2). The genome cloning and sequencing strategy for ZABYV is shown in Fig. 1B. RACE was performed using a SMARTer® RACE 5’/3’ Kit (Clontech, Dalian, China) according to the manufacturer’s instructions. The cDNAs, obtained from the RACE experiments were used as templates to amplify the middle sequences of the genome, using 2 × Phanta Max Master Mix (Vazyme, Nanjing, China). All PCR products were cloned into the pTOPO vector and sequenced using the Sanger method. The resulting sequences were assembled into a complete genomic sequence of ZABYV by DNAMAN (Version 8; Lynnon LLC, San Ramon, CA, USA), and the final sequence was 5,792 nucleotides in length. The ORFs of ZABYV, predicted using the NCBI ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/), was consistent with the genome organization of a polerovirus (Fig. 1B). In addition untranslated regions at the 5’ terminus (nt 1-130) and 3’ terminus (nt 5646-5792) and an intergenic region (nt 3411-3482), the ZABYV genome contained seven ORFs: ORF0 (nt 131-880), ORF1/ORF2 (nt 273-3400), ORF1 (nt 273-2126), ORF3a (nt 3483-3628), ORF3/ORF5 (nt 3601-5646), ORF3 (nt 3601-4203) and ORF4 (nt 3622-4207). The complete sequence and annotation of the ZABYV genome have been deposited in the GenBank database under the accession no. MK050791.
Multiple sequence alignments using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) showed that the ZABYV genome shared 45.5-48.2% nucleotide sequence identity with representative members of the genera Luteovirus and Enamovirus, and 52.5-66.6% with other documented members of the genus Polerovirus, with TuYV (66.6%), brassica yellows virus (BrYV; 65.1%) and CpCSV (63.7%) as the top three closest viruses. The same analysis using amino acid sequences translated from each ORF of ZABYV yielded sequence identity values of 14.2-31.2% in ORF0 (P0), 30.4-58.4% in ORF1 (P1), 28.3-51.5% in ORF1/ORF2 (RDRP), 37.8-77.8% in ORF3a (P3a), 40.3-94.5% in ORF3 (CP), 24.5-89.1% in ORF4 (MP) and 30.7-69.4% in ORF3/ORF5 (P3-P5). (Supplementary Table 4). These results confirmed that ZABYV is a new member of the genus Polerovirus. It is interesting that not all ORFs of ZABYV shared the highest identity with the same virus of the genus Polerovirus. The P0 protein of ZABYV had the highest sequence similarity to beet chlorosis virus (BchV; GenBank accession no. NC_002766), the P1 and RDRP had the highest sequence similarity to BrYV (GenBank accession no. NC_016038), and the other proteins (P3a, P3, P4 and P3-P5) had the highest sequence similarity to CpCSV (GenBank accession no. NC_008249), suggesting that ZABYV could be a recombinant virus (Supplementary Table 4). To determine the evolutionary relationship of ZABYV to other members of the genus Polerovirus, phylogenetic trees were constructed, using MEGA X [5], by the neighbor-joining method with 1000 bootstrap replicates based on the complete genome sequences (Fig. 1C, left) and the amino acid sequences encoded by each of the ORFs. In the phylogenetic trees based on the genome sequence and the amino acid sequences of P1 (data not shown) and RDRP (Fig. 1C, middle), ZABYV clustered closely with TuYV and BrYV, while, in the tree based on amino acid sequence of P0 (data not shown), ZABYV clustered closely with BChV, and in the trees based on amino acid sequences of P3a (data not shown), CP (Fig. 1C, right), MP (data not shown), and P3-P5 (data not shown), ZABYV clustered closely with CpCSV. The results of these phylogenetic analyses were consistent with those of multiple sequence alignments.
To find evidence of recombination events between ZABYV and other members of the genus Polerovirus, we analyzed the genome sequences of ZABYV and 19 other known poleroviruses using RDP4 [6], which includes seven recombinant detection algorithms: RDP, GENECONV, BoosScan, MaxChi, Chimaera, SiScan, and 3Seq. When aligning the genome sequence of ZABYV with those of BrYV and CpCSV, a very strong recombinant signal was detected in the region spanning nt 3,576-4,966 (Fig. 1B). In this region, the nucleotide sequence of ZABYV was 89.9% and 58.8% identical to those of CpCSV and BrYV, respectively, while in other regions, it was 52.3% and 63.9% identical, respectively (Fig. 1D). This putative recombinant event was identified by all seven algorithms in RDP4. The same signal was found in the same region of the ZABYV, TuYV and CpCSV genome sequences.
In conclusion, we have identified a new member of the genus Polerovirus, ZABYV, in zucchini grown for seed production in Xinjiang Uygur Autonomous Region, China. The complete genome sequence of ZABYV was determined using high-throughput transcriptome sequencing, overlapping RT-PCR, and RACE. Except for ORF3 and ORF4, all of the ORFs of ZABYV had less than 80% amino acid sequence identity to those of other poleroviruses. Since the amino acid sequences of all gene products of ZABYV differed by more than 10% from those of all known polerovirus, which is a criterion used to demarcate the species of the genus Polerovirus [2], we conclude that ZABYV is a new member of the genus. Based on the results of multiple sequence alignments, phylogenetic analysis, and recombination analysis, ZABYV appears to be a recombinant virus that was possibly derived from CpCSV and another novel virus, which could have a close evolutionary relationship to BrYV and TuYV. In the future, further characterization of ZABYV is needed to understand its biology and its interaction with its host.
References
Ibaba JD, Laing MD, Gubba A (2016) Pepo aphid-borne yellows virus: a new species in the genus Polerovirus. Virus Genes 53:1–3
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2011) Virus taxonomy: Ninth report of the international committee on taxonomy of viruse. Elsevier Academic, Amsterdam
Knierim D, Deng TC, Tsai WS, Green SK, Kenyon L (2010) Molecular identification of three distinct Polerovirus species and a recombinant Cucurbit aphid-borne yellows virus strain infecting cucurbit crops in Taiwan. Plant Pathol 59:991–1002
Knierim D, Maiss E, Kenyon L, Winter S, Menzel W (2015) First full-length genome sequence of the polerovirus luffa aphid-borne yellows virus (LABYV) reveals the presence of at least two consensus sequences in an isolate from Thailand. Arch Virol 160:2633–2636
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:1–5
Montero-Pau J, Blanca J, Bombarely A, Ziarsolo P, Esteras C, Martí-Gómez C, Ferriol M, Gómez P, Jamilena M, Mueller L (2018) De novo assembly of the zucchini genome reveals a whole-genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol J 16:1161–1171
Peng B, Liu LM, Liu SS, Zhang YH, Ji YL, Wu Y, Zhang XJ, Gu QS (2017) The genome of Cucurbit chlorotic yellows virus from Xinjiang autonomous region of China. Acta Phytopathol Sin 47:730–737
Ren C, Hao X, Du Y, Xiang B (2016) Molecular detection of seed zucchini virus disease in Shihezi and Xinhu region in Xinjiang. Northern Hort 40:102–106
Shang QX, Xiang HY, Han CG, Li DW, Yu JL (2009) Distribution and molecular diversity of three cucurbit-infecting poleroviruses in China. Virus Res 145:341–346
Wu Y, Li J, Peng B, Gu Q (2017) Detection of virus infecting watermelon and melon in Henan, Gansu provinces and Xinjiang Autonomous Region. China Veg 8:31–35
Xiang HY, Shang QX, Han CG, Li DW, Yu JL (2008) Complete sequence analysis reveals two distinct poleroviruses infecting cucurbits in China. Arch Virol 153:1155–1160
Zheng Y, Gao S, Padmanabhan C, Li R, Galvez M, Gutierrez D, Fuentes S, Ling KS, Kreuze J, Fei Z (2017) VirusDetect: an automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 500:130–138
Acknowledgements
We would like to thank Dr. Xuejun Zhang for assistance with the collection of zucchini samples, and Yang Wu and Suyun Qiao for technical assistance. This work was supported by Modern Agro-Industry Technology Research System (CARS-26-13), the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2015-ZFRI) and the Basic and Advanced Technology Program of Henan Province (152300410138).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Peng, B., Kang, B., Wu, H. et al. Detection and genome characterization of a novel member of the genus Polerovirus from zucchini (Cucurbita pepo) in China. Arch Virol 164, 2187–2191 (2019). https://doi.org/10.1007/s00705-019-04217-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04217-w