Abstract
Between 2010 and 2016, six mortality events were observed in Florida pompano (Trachinotus carolinus) maricultured in the Dominican Republic. Histopathological examination and conventional PCR confirmed a megalocytivirus (MCV) infection in each case. Subsequently, next-generation sequencing and phylogenomic analyses confirmed that MCV DNA was present in the infected pompano tissue samples from 2010, 2014, and 2016, and each was determined to be red seabream iridovirus (RSIV). Annotation of the RSIV genome sequences identified 121 open reading frames, and BLASTN analysis revealed the highest nucleotide sequence identity (> 99%) to a RSIV clade 1 MCV isolated from a moribund red seabream (Pagrus major) maricultured in Japan. These cases represent the first fully sequenced RSIV genomes detected outside of Asia and are the earliest reports of MCV infections in Florida pompano. This recent geographical expansion of RSIV warrants further attention to determine its potential economic and ecological impact.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus Megalocytivirus is one of three genera within the subfamily Alphairidovirinae of the family Iridoviridae [1]. Megalocytiviruses (MCVs) are double-stranded DNA viruses that continue to cause significant economic losses to both ornamental and food fish industries. The sole MCV species, Infectious spleen and kidney necrosis virus, includes three genotypes: infectious spleen and kidney necrosis virus (ISKNV), red seabream iridovirus (RSIV), and turbot reddish body iridovirus (TRBIV) [2]. Red seabream iridoviral disease (RSIVD), caused by genotypes RSIV or ISKNV, is listed as one of ten reportable fish diseases by the World Organization for Animal Health (OIE) [3]. In this study, we provide the first complete RSIV genome sequences detected outside of Asia, expanding the geographic range of RSIV into the Caribbean Sea and in a novel fish host, Florida pompano (Trachinotus carolinus).
In 2010, 2013, 2014, and 2016, a total of six mortality events occurred in Florida pompano maricultured in the Dominican Republic (Table 1). For each of the 2010, 2014a, and 2016 events, DNA was extracted from frozen spleen or kidney tissue samples using a DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer’s instructions. For the 2013, 2014b, and 2014c events, DNA was extracted from formalin-fixed paraffin-embedded tissue samples (liver, kidney, spleen, heart, intestine, eye) following a previously described protocol [2]. Histopathologic examination performed in four of the mortality events revealed cytomegalic cells with basophilic, granular, intracytoplasmic inclusions in one or more organs (liver, kidney, spleen, heart, intestine, eye), compatible with a MCV infection (Table 1). Tissue samples from all six pompano tested positive for MCV, using two specific conventional PCR assays (primers MCV-F/R and primers RSIV1-F/R) [2, 4] (Table 1). Amplified products of the expected sizes were purified using a QIAquick PCR Purification Kit (QIAGEN) and sequenced in both directions by Sanger sequencing using the aforementioned primers. BLASTN searches of the sequences revealed the highest nucleotide sequence identity (> 99%) to a RSIV genomic sequence (GenBank accession no. AB104413; Table 1) determined from moribund red seabream (Pagrus major) in Japan [5]. Hereafter, the MCVs detected in Florida pompano will be referred to as pompano iridovirus (PIV).
DNA libraries were built using a NEBNext Ultra II DNA Kit for the PIV2010 DNA sample and an Illumina TruSeq DNA PCR-Free Kit for the PIV2014a and PIV2016 DNA samples (Table S1). All three DNA libraries were sequenced using a v3 chemistry 600-cycle Kit on a MiSeq sequencer (Illumina). De novo assemblies of the paired-end reads were performed in SPAdes 3.10.1 [6] using default settings. The integrity of the complete genome sequences were verified by mapping the reads using Bowtie 2 [7] and inspecting the alignments in Tablet 1.17.08.17 [8]. Genomes PIV2010, PIV2014a, and PIV2016 were 112,321, 112,377 and 112,052 bp in size with an average coverage of 1371, 136, and 43 reads/nucleotide, respectively.
Open reading frames (ORFs) for PIV2010 (GenBank accession no. MK098185), PIV2014a (GenBank accession no. MK098186), and PIV2016 (GenBank accession no. MK098187) were predicted using the Genome Annotation Transfer Utility (GATU) with RSIV strain Ehime-1 (GenBank accession no. AB104413) as the reference genome [9]. A total of 121 ORFs were annotated for the three genomes, and the gene functions were predicted based on BLASTP searches against the National Center for Biotechnology Information (NCBI) GenBank non-redundant protein sequence database and NCBI conserved domains database.
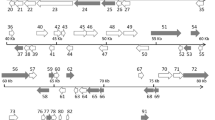
A phylogenomic analysis was performed to determine the relationship of the PIV (PIV2010, PIV2014a, and PIV2016) genomic sequences to 11 other previously sequenced MCVs. The 14 MCV genomes were aligned in Mauve 2.4.0 [10], and the resulting locally collinear blocks were concatenated using Geneious R10 [11]. A maximum-likelihood phylogenetic analysis was conducted using IQ-TREE 1.5.6 with the Bayesian information criterion option to determine the best model fit and 1000 bootstraps to determine clade support [12]. The analysis generated a well-resolved and supported phylogeny with all three PIV genomes grouping within RSIV clade 1 (Fig. 1).
Maximum-likelihood phylogram illustrating the relationship of PIV2010, PIV2014a, and PIV2016 (indicated with asterisks) to 11 other megalocytiviruses based on their aligned genome sequences. Bootstrap values are given at each node, and the branch lengths represent the number of inferred substitutions as indicated by the scale. Refer to Table S1 for virus abbreviations
The first well-characterized case of MCV disease, caused by a RSIV clade 1 MCV, occurred in red seabream cultured in Shikoku Island, Japan, in 1990 [5]. Since that time, there have been numerous reported cases of RSIV infection in marine fish species from China, Japan, Singapore, South Korea, Taiwan, and Thailand [13,14,15,16]. RSIV infection is currently considered reportable by the OIE, and a vaccine is commercially available for use in Japan [17]. Our study represents the earliest detection of a RSIV clade 1 MCV outside of Asia. Although it was beyond the scope of the current study to determine the role of PIV in the Florida pompano mortality events in the Dominican Republic reported here, our cases bear a striking resemblance to a previous report of RSIV clade 1 infection in Florida pompano associated with a mortality event in a Central American mariculture facility [18]. Florida pompano are an economically important commercial and sport fish for the United States, where the price per pound is high due to their limited supply [19]. Future studies are needed to determine the role of PIV in disease and any negative economic impact it has on the mariculture of Florida pompano in the Caribbean Sea.
References
Chinchar VG, Hick P, Ince IA, Jancovich JK, Marschang R, Qin Q, Subramaniam K, Waltzek TB, Whittington R, Williams T, Zhang Q, ICTV Report Consortium (2017) ICTV virus taxonomy profile: Iridoviridae. J Gen Virol 98:890–891
Koda SA, Subramaniam K, Francis-Floyd R, Yanong RP, Frasca S Jr, Groff JM, Popov VL, Fraser WA, Yan A, Mohan S, Waltzek TB (2018) Phylogenomic characterization of two novel members of the genus Megalocytivirus from archived ornamental fish samples. Dis Aquat Org 130:11–24
Nakajima K (2016) Chapter 2.3.8. Red seabream iridoviral disease. In: Manual of diagnostic tests for aquatic animals (World Organisation for Animal Health), 7th edn. http://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/chapitre_rsbid.pdf. Accessed 10 Oct 2018
Kurita J, Nakajima K, Hirono I, Aoki T (1998) Polymerase chain reaction (PCR) amplification of DNA of red sea bream iridovirus (RSIV). Fish Pathol 33:17–23
Inouye K, Yamano K, Maeno Y, Nakajima K, Matsuoka M, Wada Y, Sorimachi M (1992) Iridovirus infection of cultured red sea bream, Pagrus major. Fish Pathol 27:19–27
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D (2010) Tablet—next generation sequence assembly visualization. Bioinformatics 26:401–402
Tcherepanov V, Ehlers A, Upton C (2006) Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genom 7:150
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 32:268–274
Do JW, Cha SJ, Kim JS, An EJ, Park MS, Kim JW, Kim YC, Park MA, Park JW (2005) Sequence variation in the gene encoding the major capsid protein of Korean fish iridoviruses. Arch Virol 150:351–359
Huang SM, Tu C, Tseng CH, Huang CC, Chou CC, Kuo HC, Chang SK (2011) Genetic analysis of fish iridoviruses isolated in Taiwan during 2001–2009. Arch Virol 156:1505–1515
Sudthongkong C, Miyata M, Miyazaki T (2002) Viral DNA sequences of genes encoding the ATPase and the major capsid protein of tropical iridovirus isolates which are pathogenic to fishes in Japan, South China Sea and Southeast Asian countries. Arch Virol 147:2089–2109
Nakajima K, Kurita J (2005) Red sea bream iridoviral disease. Uirusu 55:115–126
Nakajima K, Maeno Y, Kurita J, Inui Y (1997) Vaccination against red sea bream iridoviral disease in red sea bream. Fish Pathol 32:205–209
Lopez-Porras A, Morales JA, Alvarado G, Koda SA, Camus A, Subramaniam K, Waltzek TB, Soto E (2018) Red seabream iridovirus associated with cultured Florida pompano Trachinotus carolinus mortality in Central America. Dis Aquat Org 130:109–115
FAO (2016) Cultured Aquatic Species Information Programme. Trachinotus spp. Text by McMaster, M.F. and Gopakumar, G. In: FAO Fisheries and Aquaculture Department, Rome. http://www.fao.org/fishery/culturedspecies/Trachinotus_spp/en. Accessed 01 Oct 2018
Acknowledgements
We thank Dr. Salvatore Frasca Jr. for his assistance in the histopathological examination and for critically reviewing the manuscript.
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All of the authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koda, S.A., Subramaniam, K., Pouder, D.B. et al. Phylogenomic characterization of red seabream iridovirus from Florida pompano Trachinotus carolinus maricultured in the Caribbean Sea. Arch Virol 164, 1209–1212 (2019). https://doi.org/10.1007/s00705-019-04155-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04155-7