Abstract
The emergence of swine-origin pandemic H1N1 (pH1N1) in 2009 invigorated extensive surveillance programs worldwide which have resulted in the deposition of large numbers of H1N1 sequences to Genbank. In the present study, we report on global evolution and dynamics of the pandemic H1N1 influenza Hemagglutinin (HA) protein in viruses isolated from three different continents (North America, Europe and Asia) during the period between April 2009 until April 2017. Close to 2000 HA full protein sequences were downloaded from the Influenza Research Database of the NCBI and analyzed using DNAStar to run an alignment, the web-based NetNglyc to predict N-Glycosylation sites and finally, the BEAST software package to calculate evolution and substitution rates. Our analysis improves upon other published papers in that we report on frequencies, dynamics and impact of HA mutations in pH1N1 viruses isolated from three continents during the past decade, as well as the evolution rate and site-specific selection pressures. Sequence based analysis demonstrated substantial changes in the HA protein over the last decade. Results showed that the HA gene is under negative selection (P value; HA= -2.253). The evolution rates varied among the three continents ranging from 2.36 × 10−3 in Europe to 3.18 × 10−3 in Asia. Mutations were detected at higher frequency and faster rate at the antigenic sites surrounding the receptor-binding domain (RBD), in particular, in the Sa and Sb sites. Mutations were either gradually accumulated to become fixed in currently circulating strains (D114N, S179N, S202T, S220T, I233T, K300E and E391K) or dynamic in terms of appearance and disappearance, both spatially and temporally (A203T, N458K and E508G). Some of the reported mutations have been shown to increase infection severity (D239G/N; globular head), enhance HA binding affinity to its receptor (S200P and S202T; RBD), or have deleterious effect on HA function (N458K and E508G; stem region). The continuous accumulation of mutations at the Sa site led to the gradual acquisition of glycosylation at residue 179 starting from 2015, which became a dominant feature in all strains isolated in the following years. In addition to sharing common amino acid substitutions (e.g. S179N in HA head and E516K in HA stem) with previous seasonal strains, the pattern of glycosylation acquisition/loss at 177 and 179 positions on the globular head, which are prominent features of immune escape, implicate that pH1N1 might follow a similar evolution trend as the SC1918 pandemic virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Influenza is the leading cause of respiratory illness worldwide. According to CDC estimations, influenza has resulted in between 9.2 million and 60.8 million illnesses and between 12,000 and 56,000 deaths annually since 2010 [1]. Three major pandemics were reported in the last century: the 1918 Spanish influenza caused by avian-origin H1N1 (SC1918) virus which killed around 20 million people [2], the 1957 Asian influenza caused by reassortant H2N2 virus, and the 1968 Hong Kong influenza caused by reassortant H3N2 virus. In April 2009, a novel H1N1 virus of swine origin was detected in humans and quickly became pandemic and resulted in more than 600,000 laboratory confirmed cases and over 18,449 deaths from its appearance until the pandemic was declared over in August 2010 [3]. Currently, the 2009 H1N1 virus is circulating, along with the predominating H3N2 virus [4].
Taxonomically, influenza A virus belongs to the Orthomyxoviridae family. Viruses harbor eight negative sense single-stranded RNA segments encoding at least 11 proteins including the two surface glycoproteins: Hemagglutinin (HA) and Neuraminidase (NA) which play pivotal roles during virus entry and release from host cells, respectively [5]. The HA glycoprotein consists of two main subunits: HA1 (327 residues) which forms the globular head and HA2 (222 residues) which constitutes the major part of the stem region [6]. The HA1 subunit carries the receptor binding domain (RBD) with five distinct antigenic sites (Sa, Sb, Ca1, Ca2 and Cb), which are the main targets of neutralizing antibodies [7]. As such, monitoring antigenic properties and amino acid changes in/around antigenic sites is necessary for the proper selection of strains recommended for use in the influenza vaccine [8]. More importantly, amino acid changes in in the RBD can alter influenza binding preference and therefore affect the inter- and intra-transmission of the virus [9].
Antigenic sites of the 2009 and 1918 HAs share a significant number of amino acid residues, especially, Sa and Sb which are located near the receptor binding site (RBS) [10]. In contrast, antigenic sites of seasonal influenza (circulating before 2009) strains show more sequence variability compared to the pandemic viruses due to several decades of virus evolution under continued immune pressure [10]. In addition to the continuous changes of amino acid sequence, or antigenic drift, seasonal influenza viruses undergo changes in the structure of HA such as the addition/removal of glycosilations. Glycosylation can occur both on the globular head and stem regions, however, these oligosaccharides sites are largely variable when found in the globular head region of HA1 and more conserved in the stem region [11]. Glycosylations on the globular head of HA play an important role in shielding antigenic sites from neutralizing antibodies; however, the acquisition of glycosylations near the receptor binding site might influence HA binding to host cell receptors and activate innate immune responses [12]. Therefore, a fine balance must be maintained regarding the optimal number and/or position of glycosylations to enable evasion of antibodies while allowing infectivity and transmission of the virus [13].
Pandemic influenza viruses express few N-linked glycosylations on the head of HA when they first appear in the human population; however, they acquire additional glycosylations as they circulate in humans. Early H3N2 strains (1968–1972), for example, harbored two potential glycosylations on the head of HA compared to six glycosylation in strains isolated later in 1980–2012 [14]. Isolates associated with the 1918 H1N1 Spanish pandemic virus expressed one glycosylation (N104), while by the 1940s most strains acquired additional glycosylations at three sites: 144, 179 and 286 [15]. When it first appeared in 2009, the swine-origin H1N1 expressed a single glycosylation site (N104) at the side of HA1 head similar to the 1918 virus, although the classical swine H1N1 virus appeared early in the 1900s and was circulating in pigs until recently [10].
Considering all the above, it is very critical to monitor influenza viruses for any emerging variants with altered pathogenicity and transmission. Several studies have been done worldwide to study the epidemiology, pathogenesis and transmission of the 2009 pandemic H1N1 virus (pH1N1). This created an extensive H1N1 sequence database [16]. Herein, we describe temporal sequence changes in the HA protein of the pH1N1 virus isolated between 2009 to 2017. Close to 2000 HA sequences (analyzed as three sets) were downloaded from the Influenza research database and aligned using ClustalW Algorithm to identify mutations and their frequencies. Sequences were also analyzed using PROVEAN to determine the potential functional consequences of the amino acid substitutions, NetNglyc server to predict potential glycosylation and MEGA7 to build the phylogenetic tree. Our data indicate rapid evolution of the HA sequence over the last decade and suggest a similar trend of evolution as that seen for the 1918 pandemic virus.
Methods
HA sequences
For this study, we selected well-identified sequences for the analysis. Preference was given to records with the full ‘month-day-year’ labels. For each of the past nine years, we chose sequences representing all months, although as expected, most cases were isolated during the winter season (October until February). In total, 1800 sequences were analyzed in three sets: Initial analysis on 900 sequences, followed by two verification tests using two sets of 450 sequences. All these sequences were from viruses isolated after April 2009. In the initial analysis, the sequences were distributed between three continents: North America (n = 472), Europe (n = 189) and Asia (n = 239). In addition, 31 HA sequences representing periods before the emergence of the 2009 pandemic were also downloaded from NCBI [17]: one sequence representative of the 1918 pandemic virus SC1918 (A/ South Carolina/1/1918), ten sequences representative of the 1977-1985 period and twenty sequences representative of the 1986-2008 period. H1 numbering was applied throughout the analysis. To confirm our findings, a similar analysis as above was done on two additional sets of 450 sequences - results are presented in the supplemental data.
Sequence alignment and phylogenetic analysis
Data were compiled and edited using the DNASTAR Lasergene sequence analysis software (DNASTAR Inc. version 13.0) and a multiple sequence alignment was conducted using ClustalW Algorithm to identify mutations in different positions of the HA protein. Phylogenetic trees were constructed with BEAST software package v1.8.4 using the maximum likelihood analysis method HKY, and gamma distribution. The substitution of amino acid sequences was defined in comparison to vaccine strains A/California/07/2009 and A/Michigan/45/2015.
Prediction of mutations’ effect on HA function
We used the PROVEAN software tool to predict the impact of amino acid substitution on the biological function of the HA protein (provean.jcvi.org) [18]. This program determines the degree of amino acid conservation in comparison to other sequences and gives a score for the potential effect of the variant on protein function. It has been used for interpreting the impact of amino acid substitutions in influenza proteins. A default cutoff score of less than − 2.5 indicates a high probability of deleterious mutations than can affect protein function [19].
Prediction of glycosylation sites
The web-based NetNglyc server was used to predict N-Glycosylation sites on the HA1 protein (cbs.dtu.dk) [20] using default settings; however, we excluded N-P-S/T sequons. Only scores crossing the default threshold of 0.5 were considered positive for potential glycosylation sites [21]. Finally, the intensive modelling Phyre2 server was used to predict and build a 3D model of the HA protein based on advanced remote homology detection methods (sbg.bio.ic.ac.uk/phyre2) [22]. The HA 3D structure of A/California/2009/04 (CA09) H1N1 (PDB 3AL4) was downloaded from the Influenza Research Database (NCBI) and was used as an input for CLC Sequence Viewer (version7-Qiagen) to manipulate the HA structure and locate antigenic sites, glycosylation sites and variant locations, relative to the different HA subunits.
Estimation of evolution rate and selection pressure
The evolution rate of the HA gene was estimated using the Bayesian Markov Chain Monte Carlo (MCMC) method as implemented in the Bayesian Evolutionary Analysis Sampling Trees (BEAST) program version v1.8.4 (http://beast.bio.ed.ac.uk) [23]. Dates of virus isolation were used to calibrate the molecular clock. The HKY substitution model was used with a gamma parameter of site heterogeneity model. A strict molecular clock model was used and evaluated by estimating the marginal likelihood implemented in the Tracer program v1.6.0 [24]. Finally, for each analysis, a chain length of 10,000,000 to 20,000,000 was used (based on number of sequences) and echoed every 1,000 states. Data uncertainty was shown in the lower and upper bound of the highest probability density (HPD) values, where 95 % of the sampled values were located. To test the significance of evolution rates among the three continents, One-way ANOVA analysis (GraphPad7) was used.
HA gene- and site-specific selection pressures for the HA gene were estimated as the ratio of non-synonymous (dN) to synonymous (dS) nucleotide substitutions per site. HA gene selective pressure detection was estimated using the Tajima test of neutrality implemented in MEGA7. Both single-likelihood ancestor counting (SLAC) and fixed-effects likelihood (FEL) methods were used to estimate dN/dS ratios at the codon level and to identify codon sites under diversifying positive selection. All these methods are available at the DataMonkey online version of the HYPhy package (http://www.datamonkey.org) [24]. In all cases, positively selected sites were defined as a dN/dS p-value less than 0.05 using the substitution model selected by the website.
Results
Intermittent and gradual acquisition of N-Glycosylations on the HA head
In 2009, circulating pandemic H1N1 strains expressed a single glycosylation at residue N104 of the HA globular head [15]. We used the online NetNglyc server to predict the presence of glycosylations on the HA protein of viruses isolated between 2009-2017 and from three continents. The N-glycosylation sequon (N-X-T/S) was detected at six sites which are conserved in almost all H1 viruses regardless of their origin; residues 28, 40, 304, 498 and 557 of the stem, and residue 104 at the side of the globular head (Figure 1). All analyzed sequences were carrying the glycosylation sequon at residues 28, 40, 304, 498 and 557. Sequence analysis revealed the acquisition of additional glycosylation site on the top of the RBD at residue 179 (within the Sa antigenic site) (Figure 1) in strains isolated in United States during 2010-2011 with a frequency of 21.7%, then in Asia in 2011 (less than 2%) before suddenly disappearing in 2012. Later in 2015, the same glycosylation sequon at position 179 was detected again in sequences isolated from USA, Asia and Europe (55%) until gradually being found in almost all sequences (90.4%) during 2016-2017 (Figure 2). In addition to the six conserved glycosylation sites, seasonal H1N1 viruses circulating prior to the 2009 pandemic virus expressed two additional glycosylation at residues 177 and either 142 or 144 of the globular head of HA protein (Table 1).
Glycosylation patterns of the HA protein of H1N1 viruses isolated after April 2009. The diagram represents the space-fill model of the H1-HA monomer (PDB 3AL4) with potential N-glycosylation in viruses isolated after April 2009. Five of these glycosylation sites (N28, N40, N104, N304 and N498) are conserved in almost all H1N1 sequences isolated since 2009 (indicated in yellow); however, the N179 glycosylation located on the top of HA1 globular head (indicated in red) became dominant in strains isolated after 2015. A few isolates from Asia had the N177 glycosylation
Dynamics of N179 glycosylation in pH1N1 isolates from North America, Asia and Europe over the nine-year period. Bars represents the total number of HA sequences analysed, black bars represents the number of sequences with potential N-glycosylation at 179 residue and dashed black line indicates the percentage of HA sequences with potential 179 glycosylation
An additional glycosylation sequon at residue 177 of HA was observed in a few strains isolated from Thailand (2009-2012) and China (2011) (Table 1 and Figure 3). An HA sequence reported from Thailand in 2009 had a similar glycosylation pattern to seasonal strains with three glycosylation sequons at 71, 142 and 177 (Table 1). Blast search for this sequence indicated 99% similarity to seasonal strains isolated in 2008 and 2009. Moreover, three HA sequences reported in Iran, one in 2014 and two in 2016, were found to possess glycosylation sequons at positions 286 and 489 while lacking glycosylation at positions 104 and 179 (Table 1). Although glycosylation at position 286 has been reported in seasonal strains isolated after 1977, N489 was not detected before 2009. Blast search for these three HA sequences revealed more than 95% similarity to A/Puerto Rico/8/1934 virus rather than the pandemic 2009 virus. Finally, one HA sequence isolated in 2010 from Switzerland showed more than 99% similarity to swine H1N1 viruses isolated during the period 2004-2012 and hence, had a different glycosylation pattern than human H1N1 virus. This virus lacked the glycosylations at positions 104, 179 and 304 but expressed a glycosylation at position 291 of the HA protein (Table 1).
Structure of the HA monomer showing antigenic sites and common amino acid substitutions detected in these sites between 2009 and 2017. Five antigenic sites are located at the globular head of the HA1 subunit, Sa (Fuentes-Gonzalez et al.), Sb (yellow), Ca1 (purple), Ca2 (blue) and Cb (green). With the exception of D239 and A203T, the mentioned mutations have been reported in all sequences detected in 2017 (predominant variants). Substitutions at the Sa site (S179 and K180) resulted in the acquisition of a new glycosylation site at 179–an Asparagine residue on the top of the HA globular head in all sequences isolated after 2016
Amino acid substitutions at the RBD-A region and antigenic sites
The top part of the RBD near the sialic acid recognition site has a large subdomain referred to as RBD-A. RBD-A has been previously shown by Wei and colleagues (2010) to show 95% similarity between two the pandemic strains of SC1918 and CA09 [15] . Sequence analysis indicated that the RBD-A region has undergone dramatic changes since the emergence of pandemic 2009 H1N1 virus. By the end of 2009, the RBD-A region was already showing 10% difference with the original A/California/04/2009 H1N1 strain (CA09). By 2016, sequence identity in the RBD-A region had dropped to 77%, between the circulating strains and the CA09 isolate (Table 2).
Comparison of HA antigenic sites between pandemic and seasonal strains is shown in Table 3. As has been reported earlier [10], the CA09 and SC1918 H1N1 pandemic viruses share a significant number of amino acid residues in their antigenic sites (83%) (Table 3). Only one amino substitution was observed in antigenic site Sa (92% similarity) between the two viruses. Also, only two amino acids mutations were reported in the Sb site of pandemic CA09 as compared to the SC1918 pandemic strain (83% similarity). The Ca site, on the other hand, showed 63% similarity between pandemic viruses, and no substitutions were detected in the Cb site. Recent isolates from 2016 had limited and conserved amino acid changes compared to the CA09 pandemic virus including: S179N and K180Q in the Sa site, S202T in the Sb site, and I183V and R222K in the Ca site.
Evolution rate and selective pressure
The average overall mean evolution rate of the H1 gene of the pH1N1 viruses analysed in this study amounted to 2.83 × 10−3 with the highest rate reported in Asia (3.18 × 10−3) followed by North America with 2.94 × 10−3 and lastly Europe with 2.36 × 10−3 (Table 4). However, no significant differences were found among the three continents using an One-Way ANOVA test. Assessing the dN/dS ratio at the gene level showed that the HA gene is under negative selection with a Tajima’s D value of -2.253 (positive selection is considered when a P value of dN/dS ratio is > 1). At the site level, however, five codons; two in the signal peptide at positions 2 and 3, and three codons in the coding region (100, 128, 179) were identified to be under diversifying positive selection when applying the FEL method on HA sequences of pH1N1 viruses isolated from North America (Table 4).
Mutation types, dynamics and impact on HA biological function
Most of the detected mutations, which are characterized by the complete replacement of amino acids, were conserved and neutral (Table 5). Among these, mutations D114N, S202T, S220T, K300E, E391K and E516K were the earliest to be detected and were divergent from pandemic CA09 virus within three years of its emergence. Almost all circulating viruses had a S220T substitution at Ca1 antigenic site by the end of 2009 (Table 5). The D114N change was observed at a lower rate and was first detected in the USA during September of 2009 (frequency of 7.5%) and later in Asia (11%) and Europe (22%) in 2010. Complete replacement at position 114 (D to N) was observed globally in 2013. Mutations at the Sa antigenic site, in particular S179N, led to the glycosylation acquisition in viruses isolated since 2015. Additionally, the K180Q mutation in the Sa site accumulated rapidly since its appearance in 2012 in Europe (10%) and Asia (22%) and in 2013 in North America (65%) becoming the dominant strain worldwide in 2015 (Supplementary figure 1). The K300E and A273T mutations were detected in viruses isolated in USA during 2009 (4%) and 2011 (5%), respectively. K300E was reported later in strains isolated in 2012 from Europe (30%) and Asia (33%). In 2015, all sequences isolated from USA, Europe and Asia were carrying these two substitutions. Only two substitutions were detected shortly after the 2009 pandemic in the HA2 subunit: E391K and E516K (Figure 4). E391K, which is thought to be important for membrane fusion [25], was found in 3% of sequences isolated from USA, Europe and Asia. Less than 1% of sequences were carrying E516K mutation by the end of 2009. However, the frequency of both substitutions increased rapidly until strains carrying E391K and E516K were the major circulating strains (>95%), in 2011 and 2013 respectively (Supplementary figure 1).
Mutations detected in the HA protein of pH1N1 viruses isolated from 2009 until 2017. The figure depicts mutations acquired by H1 HA as it evolved between 2009-2017. Glycosylation sites are indicated in faint red except the glycosylation at 179 site (bright red) which is found on the top of the HA1 globular domain. Detected mutations are indicated as vertical lines: Straight lines represents mutations with complete replacement while dashed lines indicates mutations found in some sequences
A few neutral variants were also found temporarily in certain geographical regions such as N48D, which was detected in 12.5% of sequences isolated from USA during 2010 and disappeared in the following years. Another substitution in the Sb region, A203T, was also reported in the USA during 2009 at very low frequency (<1%); this peaked in 2010 to reach 15% of sequences before decreasing again in 2014 (5%) and disappearing later on. This substitution was also reported in Europe during the period 2009-2014 starting with less than 1.5% frequency at 2009 and rising to reach 33% in 2015 (supplementary figure 1). S145P is another mutation that was detected first in Asia during 2009 (4%), followed by a gradual increase to reach 18% of sequences reported in 2010 before totally disappearing by 2012. N387H and N458K variants were found exclusively in Europe. The N387H change appeared by the end of 2009 in about 15% of strains and then dropped to 10% in 2010 before disappearing entirely by late 2010. Finally, the N277D variant was detected during 2012 in USA (50%) and in Europe (20%) but disappeared in later years.
Further analysis of detected variants using PROVEAN tools revealed that some of mutations have a deleterious effect on HA function, thus, explaining their rare frequency and rapid disappearance. One of these mutations is N458K, which was found in 29% of strains isolated from Europe during 2010 and then disappeared in following years. Another mutation was found at residue 508 in which a Glutamic acid was replaced by a polar amino acid, Glycine. This mutation’s PROVEAN score (−3.597) suggests a major impact on HA function. The mutation was detected during 2014 in 8.5% of USA strains and during 2016 in 17% of reported viruses in Asia. Interestingly, strains which were found to carry E508G mutation in Asia were also shown to have a neutral substitution at 518 position (D518E). Finally, we detected some rare mutations that were previously reported to affect virus binding affinity and pathogenicity, nonetheless, these mutations did not show any spatial or temporal preferences [26, 27]. An example of such mutations is D239N/E/G in the Ca2 antigenic site (Figure 4). The Aspartic acid residue at position 239 was substituted by N (0.7%), E (1%) or G (1.1%) in all sequences included in the study. Another mutation located in the Sa site, G172E, was reported in 0.66% of the analyzed sequences, including two sequences from North America in 2013 and 2017. This mutation was found to enhance virus virulence in mice [28] (Table 5).
Other observed mutations that were previously shown to affect HA functionality are A151T, S200P, S202T, T214A and I233T. While S200P and S202T were found to increase the receptor-binding avidity, A151T and A214T are known to decrease binding avidity [29]. S200P was detected first in USA in 2010 (2.5%) and gradually detected at higher frequencies in following years reaching 44% as of 2017 (Supplementary figure 1). Strains carrying this substitution were also detected during the periods 2009-2010 and 2010-2013 in Asia (2.8%) and Europe (14.1%) respectively, and disappeared afterwards. The S202T substitution in the Sb site (Figure 4) was first detected in 2010 in USA (17.5%), Europe (35%) and Asia (25%), and became the predominate variant (98%) in 2013 isolates. On the other hand, isolates with T214A substitutions were reported in Asia early during the 2009 pandemic (8.5%) and shortly afterwards in USA and Europe (supplementary figure 1). Despite its negative effect on receptor binding, this mutation rapidly increased to reach 56% prevalence in 2012 but declined thereafter and disappeared completely in 2016 from all analyzed sequences. A151T, another substitution that is thought to decrease binding avidity of HA, appeared in 2009 at low frequencies and then disappeared soon after in 2012. Interestingly, sequences expressing the A151T variant were always associated with S200P and S202T but never with A214T. Finally, we observed gradual displacement of the non-polar isoleucine with a polar amino acid threonine at residue 233 of the RBS. I233T appeared first in Europe during 2009 (5.3%) then in USA during 2012 (10.7%) until it became the major circulating variant in 2016 (Supplementary figure 2). However, I233T was not detected until 2015 in Asia (41%) and continued to be the major strain in these continents (Table 5).
To check the redundancy of our results, we run similar analysis as indicated above on two additional sets of HA protein sequences. Each set includes 150 sequences per continent (450 in total) over the nine-year period. The six analyzed variants in these two groups showed similar patterns and frequencies as reported in the initially characterized 900 sequences (Supplementary figure 2).
Phylogenetic analysis
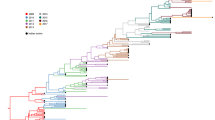
Phylogenetic analysis was performed to investigate the temporal and geographical evolution of H1N1 viruses over the past nine years (Figure 5). Analysis revealed the presence of distinct phylogenetic groups, where viruses were grouped based on the year of isolation in all three continents (Supplementary figure 3). Viruses isolated between 2009-2010 grouped in a separate clade to the rest of the viruses. Sequence variability was more obvious in strains isolated between 2010-2011. Expectedly, the vaccine strain A/California/07/2009 (H1N1) grouped with viruses sequenced during 2009 with an average similarity of 98.9% (Figure 5). Three strains were outliers, two from Iran and one from Switzerland, and these were shown not to be of 2009 pH1N1 origin.
Phylogenetic tree of HA amino acid sequences isolated during the period 2009-2017 from North America, Europe and Asia. The phylogenic tree was constructed using the maximum likelihood method by Jones-Taylor Thornton model. The red square indicates the H1N1 vaccine strain, A/California/07/2009, recommended by WHO. Three sequences were identified as outliers, two isolated from Iran during 2014 and 2016 (blue star) and one from Switzerland in 2010 (green star). BLAST analysis of these strains has shown that they show closer similarity to A/Puerto Rico/8/1934 virus (blue star) and swine H1N1 virus (green star)
Discussion
A total of 1800 (three sets) randomly selected HA sequences of H1N1 viruses isolated after April 2009 from North America, Europe and Asia were analyzed in this study. Following the H1N1 pandemic in 2009, several studies were conducted worldwide to evaluate genetic and antigenic evolution of the virus; however, this is the first study that analyzes sequences from three continents over a nine-year period, and report on mutation types, frequencies, dynamics and their possible impact on virus behavior. Overall, a series of HA mutations were identified over the study period, when compared to the original sequence of pandemic 2009 virus. Serial sequence sampling throughout the nine years revealed that HA mutations were either gradually accumulated to become stable in circulating strains (D114N, S179N, S202T, S220T, I233T, K300E and E391K) or dynamic in terms of appearance and disappearance, spatially and temporally (A203T, N458K and E508G). Although mutations were detected in different domains, most of the reported mutations were located in antigenic sites surrounding the receptor-binding site, which are the target for neutralizing antibodies [10]. Most of the reported mutations were previously shown not to affect the antigenic property of the HA protein, as determined by hemagglutination inhibition (HAI) assay [30]. This explains the consistency in the vaccine composition since 2009, whereby the H1N1 component was only changed this year after WHO recommendation. Although most of the recently isolated H1N1 viruses were shown to be antigenically indistinguishable from the original vaccine strains A/California/7/2009 (2016-2017; northern hemisphere), these viruses were poorly inhibited by some post-vaccination (A/California/7/2009) adult human sera [30]. Accordingly, the A/Michigan/45/2015 virus was recently recommended to be included in the vaccine composition for the 2017-2018 season in both hemispheres. This virus exhibit 97% similarity in the HA sequence to the original A/California/7/2009 virus. It also carries significant mutations in the antigenic sites including: S179N and K180Q in the Sa site, S202T in the Sb site and S222T in the Ca1 site.
On the other hand, a few studies were conducted to evaluate the effect of detected mutations on virus transmission and pathogenicity. Some of these mutations have been reported to affect virus binding affinity to human receptors such as D239 and D204 [27]. The presence of Aspartic acid at these positions is a critical feature in all human-adapted H1N1 viruses [27]. Experimentally, single substitution of D to G at position 239 has been shown to increase affinity for alpha 2-3 sialic acid receptors in addition to its affinity to human alpha 2-6 receptors [9]. Further, D239G/N substitutions have been detected in severe respiratory infections only, while the G239E variant was detected in both severe and mild cases at similar frequencies [26]. According to our analysis, strains carrying these variants were rarely detected spatially and temporally, with frequencies reaching 0.7% and 1.1% for D239G and G239N, respectively. The majority of sequences analyzed in this paper possess D204 and D239 (>97%).
In a review published in 2012, N. Sriwilaijaroen and Y. Suzuki predicted changes in specific amino acids within the vicinity of the RBS that would enhance HA affinity to human receptors. Specifically, mutations at residues I233, E241 and T214 that have direct interaction with S200, D204 and Q205 respectively, would change the amino acid networking within the RBS and enhance its preference to human receptor [9]. A comparison of amino acids at positions 233 and 241 across a number of HA sequences indicated that they are either both hydrophobic such as that observed in pandemic SC18 HA, or they are both charged such as in A/Brisbane/ 59/2007 [31]. In contrast, HAs of pH1N1 viruses maintain a combination of I233 and E241, amino acids that are highly conserved in strains isolated between 2009 and 2015. This mismatched combination has been proposed to disrupt the positioning of residues in the RBS and the formation of a stable network of interactions [31]. Experimental substitution of Isoleucine with Lysine at position 233 has been shown to markedly increased HA affinity to the human receptor [31]. In our study, we reported the gradual displacement of the non-polar Isoleucine residue at 233 position with a polar amino acid, Threonine, reaching 96% frequency in year 2017. This substitution could generate a stable ionic interaction between polar T233 residue in the RBS and the acidic, E241 residue in the 220 loop, consequently increasing binding affinity to the human receptor in comparison to earlier H1N1 isolates [31]. Further, as predicted by N. Sriwilaijaroen and Y. Suzuki, the threonine residue at position 214 was replaced gradually by alanine until reaching 56% frequency in 2012; however, this disappeared again in 2015. Some substitutions at the RBS have been reported to exhibit opposite individual effects on receptor binding avidity. S200P and S202T substitutions are known to increase the receptor-binding avidity of HA, whereas A151T and A214T substitutions tend to decrease binding avidity [29]. The later mutations, A151T and A214T, were detected in fewer strains and for a short period of time, which partially explain their rapid disappearance from currently circulating stains. On the other hand, the S202T and S200P variants were first detected in 2010 in all three continents. S202T became the predominate variant in 2012 isolates (>96%) while S200P mutation gradually increased in frequency over time to reach 44% in 2017 in the USA while disappearing from Asia in 2012 and from Europe in 2015. Strikingly, our analysis has shown that two major variants harboring combinations of these substitutions were detected during the 2010- 2011 period: S200P-A151T and S202T-A214T, respectively. The A151T variant, for example, was always associated with S200P (100%) but never with A214T, in comparison with previously circulating strains that had A151, S200 and S202. The coexistence of A151T and A214T could have detrimental effects on virus replication and transmission [29], which partially explains our results. The combination of these substitutions (A151T and S200P) would maintain the receptor-binding properties of the virus (Table 5).
A study that investigated the effect of the S200P mutation after its appearance in 2010 demonstrated that it can dramatically increase virulence of pH1N1 virus in mice, either alone or in combination with D144E. Interestingly, these two mutations were also reported in the 1918 and seasonal H1N1 viruses. In our analysis, only 0.8% of sequences were carrying the D144E mutation and were detected in all continents at different periods (2009-2015). Only one HA sequence was carrying both mutations, S200P and D144E, and that was in a virus isolated in Switzerland in 2010 which showed more than 99% similarity (BLAST search) to swine H1N1 virus [32].
In addition to the mutations in the RBD, we reported E391K and E516K mutations at the stem region. The most prominent mutation was E391K in the proximity of the fusion peptide which was first described in 2010 [25]. E391K was first identified in New York in July of 2009 and observed shortly afterwards in viruses isolated worldwide. According to our analysis, this mutation increased rapidly in 2010 (50%) and became the major circulating variant in 2011 with more than 96% frequency. Although E391K mutation has not been linked to severe infections, viruses with this variant have been detected in pandemic vaccine breakthrough infections [33]. The rapid expansion of E391K mutation since its early appearance in late 2009 could be explained partially by its role in forming an intermolecular interactions between HA monomers and hence increased HA trimer stability [34].
Seasonal strains (1988-2008) showed only 67% sequence identity with the pandemic 1918 SC virus in the RBD-A region [15]. Similarly, strains isolated after 2009 pandemic expressed rapid mutation rate in RBD-A reaching 77.6% by the end of 2016, however, the antigenic drift dramatically decreased after the acquisition of glycosylation at position 179 in the Sa site. (Table 2). Therefore, changes in the RBD-A region contribute largely to neutralization resistance and viral evolution in humans. Neither pandemic strains were glycosylated on the top of the HA head (Table 1). However, the seasonal strains following the 1918 pandemic expressed two highly conserved glycosylation sites (142 and 177) within the Sa antigenic site of strains isolated during 1986 and 2008, which have been shown to play a role in evading the human immune response [15]. The similar glycosylation pattern of both pandemic viruses in the HA head prompted us and others to estimate evolutionary rate and patterns in comparison with the seasonal H1N1 [35]. Our analysis indicated two separate appearances of 179 glycosylation: one in 2010 reaching 20% frequency, which rapidly disappeared in 2013, and another in 2015 that gradually increased in frequency to more than 98% in 2016 isolates (Figure 1). Interestingly, 179 glycosylation was among the first glycosylations to be reported in seasonal H1N1 influenza (1933–1949) although it was not detected in later strains (1950-2008) [15, 36]. The alterations in glycosylation sites following the 1918 H1N1 pandemic was studied extensively by Sun et al in 2011 and they reported similar patterns to what we have found in this study [36]. The glycosylation at position N179 is thought to disturb the interaction between HA-RBS and its receptor, which could partly explain its disappearance from seasonal strains isolated after 1950s. Sun et al (2012) have also proposed that the positional conversion of N179 to N177 occurred in 1951 may have been driven by a virus requirement for enhanced binding of HA to its receptor [37]. We detected very few strains with the N177 glycosylation and all were from Asia.
In contrast to our findings, a previous study carried out by Job et al in 2013 reported that the glycosylation at 179 position was prevalent in sequences isolated during 2009, and declined steadily in 2012 before disappearing totally by 2013 [12]. Further, they found that viruses expressing the N179 glycosylation are similarly sensitive to neutralization as compared to those that do not express glycosylation, suggesting that glycosylations at N179 may not play a major role in antigenic shielding in spite of its location within the Sa antigenic site [12]. Algorithms capable of predicting the timeline of IAV glycosylation evolution from the emergence of the pandemic until eventual replacement are now available and could be used for further analysis [38].
Although HA sequences were grouped per year rather than site of isolation in the phylogenetic analysis, the patterns and frequencies of certain mutations (179 glycosylation for example; figure 2) were not consistent between isolates from different continents, suggesting that viruses might follow different evolutionary patterns at different locations. This could partially depend on the overall herd immunity in the different areas, where influenza vaccines are widely used in North America, to a lesser extent in Europe, and not much in Asia.
Conclusion
In conclusion, we report here on the global evolution and dynamics of the H1N1 HA since its emergence in 2009. Our analysis improves upon other published papers in that we report on frequencies, dynamics and impact of HA mutations in viruses isolated worldwide during the past decade, as well as the evolution rate and site-specific selection pressures. As expected, the majority of mutations were detected in the antigenic sites of the head of HA domain. However, we also reported mutations at the stem region such as E391K and E516K. Few mutations were shown to increase the receptor binding affinity (S200P and S200T), others were shown to affect virulence (G172E), and most recorded mutations were found to have no significant effect on HA function with the exception of D239 at the head region and N458K and E508G at the stem region. Interestingly, one glycosylation at residue 179 was acquired after nine years of virus circulation, which was dynamic in appearance and disappearance as observed within seasonal strains. Another glycosylation at residue 177 which was also observed in the seasonal stains, was only detected in a few isolates from Asia. This glycosylation is predicted to replace the 179 glycosylation, as observed in seasonal strains that circulated between 1918 and 2009. This indicates that currently circulating H1N1 viruses might follow similar evolution patterns to those observed in seasonal H1N1 viruses that circulated before 2009. Interestingly, only a few sequences (isolated after April 2009) carried similar glycosylation and showed more than 95% similarity (BLAST search) to H1N1 viruses isolated before the pandemic, indicating that seasonal H1N1 strains might not have totally disappeared and they could be still circulating at very low frequencies.
References
Rolfes MA, Foppa IM, Garg S, Flannery B, Brammer L, Singleton JA et al (2013) Estimated influenza illnesses, medical visits, hospitalizations, and deaths averted by vaccination in the United States, 9 Dec 2016. Available from: https://www.cdc.gov/flu/about/disease/2015-16.htm. Accessed 15 Mar 2018
Johnson NP, Mueller J (2002) Updating the accounts: global mortality of the 1918-1920” Spanish” influenza pandemic. Bull Hist Med 76(1):105–115
WHO (2010) Pandemic (H1N1) 2009—update 112
WHO (2017) 21 August 2017,—Update number 296, based on data up to 06 August 2017. In: WHO report. WHO
Gamblin SJ, Skehel JJ (2010) Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285(37):28403–28409
Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P (2009) A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83(16):8021–8031
Luoh SM, McGregor MW, Hinshaw VS (1992) Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J Virol 66(2):1066–1073
World Health Organization (WHO) (2017) Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Available from: http://www.who.int/influenza/vaccines/virus/characteristics_virus_vaccines/en/. Accessed 11 Feb 2018
Sriwilaijaroen N, Suzuki Y (2012) Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci 88(6):226–249
Igarashi M, Ito K, Yoshida R, Tomabechi D, Kida H, Takada A (2010) Predicting the antigenic structure of the pandemic (H1N1) 2009 influenza virus hemagglutinin. PLoS One 5(1):e8553
Wang CC, Chen JR, Tseng YC, Hsu CH, Hung YF, Chen SW, Chen CM, Khoo KH, Cheng TJ, Cheng YS et al (2009) Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci USA 106(43):18137–18142
Job ER, Deng YM, Barfod KK, Tate MD, Caldwell N, Reddiex S, Maurer-Stroh S, Brooks AG, Reading PC (2013) Addition of glycosylation to influenza A virus hemagglutinin modulates antibody-mediated recognition of H1N1 2009 pandemic viruses. J Immunol (Baltimore, Md : 1950) 190(5):2169–2177
Schulze IT (1997) Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J Infect Dis 176(Suppl 1):S24–S28
Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B (2004) Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14(12):1229–1246
Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, Yang ZY, Tumpey TM, Nabel GJ (2010) Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med 2(24):24ra21
Influenza Research Database. https://www.fludb.org/. Accessed 20 Apr 2017
Influenza Research Database: https://www.fludb.org/. Accessed 20 April 2017
PROVEAN. http://provean.jcvi.org/. Accessed 25 Aug 2017
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10):e46688
NetNGlyc 1.0 server: http://www.cbs.dtu.dk/. Accessed 20 July 2017
Gupta R, Brunak S (2002) Prediction of glycosylation across the human proteome and the correlation to protein function. In: Pacific Symposium on Biocomputing, vol 7. Bio-Centrum Technical University of Denmark, Lyngby, pp 310–322
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Pond SL, Frost SC, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics (Oxford, England) 21(5):676–679
Maurer-Stroh S, Lee RT, Eisenhaber F, Cui L, Phuah SP, Lin RT (2010) A new common mutation in the hemagglutinin of the 2009 (H1N1) influenza A virus. PLoS Curr 2:rn1162
Valli MB, Selleri M, Meschi S, Zaccaro P, Vincenti D, Lalle E, Capobianchi MR, Menzo S (2011) Hemagglutinin 222 variants in pandemic (H1N1) 2009 virus. Emerg Infect Dis 17(4):749–751
Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM et al (2007) A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science (New York, NY) 315(5812):655–659
Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ (2010) Adaptation of pandemic H1N1 influenza viruses in mice. J Virol 84(17):8607–8616
de Vries RP, de Vries E, Martinez-Romero C, McBride R, van Kuppeveld FJ, Rottier PJ, Garcia-Sastre A, Paulson JC, de Haan CA (2013) Evolution of the hemagglutinin protein of the new pandemic H1N1 influenza virus: maintaining optimal receptor binding by compensatory substitutions. J Virol 87(24):13868–13877
World Health Organization-Vaccines (2017) WHO recommendations on the composition of influenza virus vaccines. Available from: http://www.who.int/influenza/vaccines/virus/recommendations/en/. Accessed 27 Dec 2017
Jayaraman A, Pappas C, Raman R, Belser JA, Viswanathan K, Shriver Z, Tumpey TM, Sasisekharan R (2011) A single base-pair change in 2009 H1N1 hemagglutinin increases human receptor affinity and leads to efficient airborne viral transmission in ferrets. PLoS One 6(3):e17616
Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA et al (2010) Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog 6(10):e1001145
Barr IG, Cui L, Komadina N, Lee RT, Lin RT, Deng Y, Caldwell N, Shaw R, Maurer-Stroh S (2010) A new pandemic influenza A(H1N1) genetic variant predominated in the winter 2010 influenza season in Australia, New Zealand and Singapore. Eur Surv 15(42):19692. https://doi.org/10.2807/ese.15.42.19692-en
Yang H, Chang JC, Guo Z, Carney PJ, Shore DA, Donis RO, Cox NJ, Villanueva JM, Klimov AI, Stevens J (2014) Structural stability of influenza A(H1N1)pdm09 virus hemagglutinins. J Virol 88(9):4828–4838
Kim JI, Lee I, Park S, Hwang MW, Bae JY, Lee S, Heo J, Park MS, Garcia-Sastre A, Park MS (2013) Genetic requirement for hemagglutinin glycosylation and its implications for influenza A H1N1 virus evolution. J Virol 87(13):7539–7549
Sun S, Wang Q, Zhao F, Chen W, Li Z (2011) Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One 6(7):e22844
Sun S, Wang Q, Zhao F, Chen W, Li Z (2012) Prediction of biological functions on glycosylation site migrations in human influenza H1N1 viruses. PLoS One 7(2):e32119
Altman MO, Angel M, Košík I, Gibbs JS, Trovão NS, Zost SJ, Hensley SE, Nelson MI, Yewdell JW (2017) Hemagglutinin glycan clock guides human influenza a virus evolution. bioRxiv. https://doi.org/10.1101/163345
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Availability of data and material
The datasets analysed during the current study are available in the in Influenza Research Database, https://www.fludb.org/.
Additional information
Handling Editor: Hans Dieter Klenk.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Al Khatib, H.A., Al Thani, A.A. & Yassine, H.M. Evolution and dynamics of the pandemic H1N1 influenza hemagglutinin protein from 2009 to 2017. Arch Virol 163, 3035–3049 (2018). https://doi.org/10.1007/s00705-018-3962-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3962-z