Abstract
The mycoparasitic oomycete Pythium nunn isolate UZ415 contains two double-stranded RNAs (dsRNAs) of different sizes. The 1707-nt dsRNA1 and the 1475-nt dsRNA2 potentially encode an RNA-dependent RNA polymerase (RdRp) and a coat protein (CP), respectively, with sequence similarity to the RdRp and CP of gammapartitiviruses (< 57% and < 36%). Phylogenetic analysis of the deduced RdRp amino acid sequences indicated that the virus identified from P. nunn is classifiable as a distinct member of the genus Ganmmapartitivirus in the family Partitiviridae. This virus isolate is hereby named Pythium nunn virus 1 (PnV1).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Oomycetes are fungi-like filamentous eukaryotic microorganisms that belong to a phylogenetic lineage distinct from that of fungi. Oomycetes are classified in the phylum Heterokontophyta (the “stramenopiles”) in the kingdom Chromalveolata, to which brown and golden algae and diatoms also belong. Although little has been reported on viral infections in oomycetes, some viruses have been found in the plant pathogenic oomycetes, Phytophthora infestans, Phytophthora spp., Sclerophthora macrospora, and Plasmopara halstedii [1,2,3, 5, 6, 13].
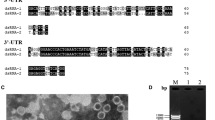
Pythium, a genus belonging to the class Oomycotes, consists of over 150 species, most of which are plant parasites and some saprophytes. Virus-like dsRNA and icosahedral virus-like particles have been found in P. irregulare [4]; however, there has not been a report on the genome sequences of the viruses in Pythium spp. During our exploration for oomycete viruses in Pythium spp., we found two dsRNA segments in an isolate of P. nunn (Fig. 1a). Pythium nunn is a mycoparasitic Pythium species that does not cause plant disease and has been studied extensively as biocontrol agent against fungi and oomycetes plant pathogens [7]. It is classified in clade I in the 11 smaller clades under Pythium [8]. Here, we report a novel partitivirus identified from P. nunn.
a: dsRNA profile of PnV1 observed through 1% agarose gel electrophoresis. Arrowheads indicate the two dsRNA segments from Pythium nunn isolate UZ415 (Pu415). M, 1 kb DNA ladder (New England Biolabs). b: Genome organization of PnV1. Each dsRNA has a single open reading frame (ORF); the ORF of one dsRNA encodes an RNA-dependent RNA polymerase (RdRp) while that of the other dsRNA encodes a capsid protein. The boxes and solid lines indicate the ORFs and untranslated regions (UTR), respectively. The UTR lengths are shown under a solid line
The P. nunn isolate UZ415, which was isolated from soils in a deciduous forest in Fukuoka Prefecture in Japan [7], was characterised. Mycelia were propagated on potato dextrose liquid medium in an autoclaved ziplock container (156 × 117 × 53 mm) at 25 °C for 7 days. Total nucleic acid was extracted from the mycelia and dsRNA was purified using CF-11 cellulose following previously described methods [10]. After digestion with RNase-free DNase I and S1 nuclease, the size and quality of the dsRNAs were assessed through 1.0% (w/v) agarose gel electrophoresis.
In preparation for deep sequencing, a library was constructed from 84 ng of the dsRNA using an NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA). Sequencing was performed on a MiSeq benchtop sequencer using a MiSeq Reagent Kit Nano V2 (300 cycles) (Illumina, San Diego, CA), with paired-end reads being obtained. De novo assembly of the trimmed-raw reads was performed using the program Velvet. Two contigs were found to be similar to the RdRp and CP genes of partitiviruses. From a total of 991,052 raw reads, the contig coding for RdRp was assembled from 917 reads while the contig coding for CP was assembled from 577 reads. After sequencing the 5′ and 3′ ends of the two viral contigs by RNA ligase-mediated amplification of cDNA ends (RLM-RACE) [9], full-length cDNA was generated from each dsRNA through RT-PCR using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), PrimeStar GXL DNA polymerase (Takara, Otsu, Japan), and 5′- and 3′-end specific primers (Table S1). No differences were found between the viral genome sequence obtained through Sanger sequencing of RT-PCR products and the sequence obtained through MiSeq sequencing of dsRNA. The complete nucleotide sequences of the two dsRNA segments were submitted to GenBank with accession numbers LC371062 and LC371063.
The complete sequences of dsRNA 1 and 2 of this putative oomycete virus isolate, which we have named Pythium nunn virus 1 (PnV1), were determined to be 1707- and 1475-nt long, respectively (Fig. 1b), with GC contents of 44.5% and 45.2%, respectively. Both dsRNAs contain a single open reading frame (ORF) with 5′ and 3′ untranslated regions (UTRs) that are 22 and 92 nt in length for dsRNA1, respectively, and 46 and 148 nt in length for dsRNA2, respectively. The 5′ termini of both dsRNA molecules contain an 8-nt long conserved sequence (CGT TGA AT).
Sequence analysis of dsRNA1 revealed that it contained a single ORF (ORF1) on its plus strand starting at nt 23 and ending at nt 1615, putatively encoding a protein with 530 amino acids. A BLASTp search showed that this protein has significant sequence similarity to the RdRps of viruses classified within the family Partitiviridae. The top 3 hits were Penicillium stoloniferum virus F (PsV-F; identity, 57%; query cover, 96%; e-value, 0), Magnaporthe oryzae partitivirus 2 (MoPV1; identity, 56%; query cover, 96%; e-value, 0), and Magnaporthe oryzae partitivirus 1 (MoPV2, identity, 55%; query cover, 96%; e-value, 0). We also found an RdRp conserved domain (cd01699, e-value = 4.85e-09) using a conserved domain database search. Multiple-protein alignment of the deduced amino acid sequences of the RdRps revealed that the ORF1 of PnV1 has six conserved motifs (III to VIII) that are present in the RdRps of partitiviruses (Supplemental Fig. 1). dsRNA2 was also found to contain a single ORF (from nt 47 to 1327, ORF2), which putatively encodes a 427-aa protein. A BLASTp search showed that the sequence of this ORF is similar to those of the coat proteins (CPs) of gammapartitiviruses, although only 3 hits were found: MoPV2 (identity, 36%; query cover, 96%; e-value, 8e-87), MoPV1 (identity, 36%; query cover, 96%; e-value, 1e-86), and PsV-F (identity, 39%; query cover, 95%; e-value, 1e-82).
A Maximum-likelihood (ML) tree was constructed from the deduced RdRp amino acid sequences of PnV1 and those of confirmed and putative viruses classified in the family Partitiviridae using MEGA 7.0. The ML tree clearly showed that PnV1 clustered with PsV-F, MoPV1, and MoPV2 in the clade gammapartitivirus (Fig. 2). The species demarcation criteria for the genus Gammapartitivirus is an RdRp amino acid sequence identity less than 90% and a CP amino acid sequence identity less than 80% [11, 12]. The amino acid sequence identity between the RdRp and CP of PnV1 and those of the virus with which it had the highest percent identity (PsV-F) was 57% and 39%, respectively. This indicates that PnV1 is an isolate representing a putative new virus species belonging to the genus Gammapartitivirus. To our knowledge, this is the first report of a partitivirus identified from an oomycete species.
Phylogenetic analysis of the deduced amino acid sequence of the RdRp of PnV1 and those of representative viruses classified in the family Partitiviridae or other partitivirus-like sequences. Amino acid sequences were aligned using clustal W, and a phylogenetic tree was constructed using the Maximum-likelihood method based on the Jones-Taylor-Thornton model with 1,000 bootstrap replicates in MEGA 7.0. Virus abbreviations and accession numbers are as follows: white clover cryptic virus 1 (WCCV1; AY705784), Rhizoctonia solani dsRNA virus 2 (RHsdRV2; KF372436), Heterobasidion partitivirus 3 (HetPV3; FJ816271), Chondrostereum purpureum cryptic virus 1 (CpCV1; AM999771), Rosellinia necatrix partitivirus 2 (RnPV2; AB569997), Sclerotinia sclerotiorum partitivirus S (SsPV-S; GQ280377), Heterobasidion partitivirus 2 (HetPV2; HM565953), Heterobasidion partitivirus 7 (HetPV7; JN606091), Atkinsonella hypoxylon virus (AhV; L39125), Ceratocystis resinifera virus 1 (CrV1; AY603052), Rhizoctonia solani virus 717 (RHsV717; AF133290), Heterobasidion partitivirus 8 (HetPV8; JX625227), Pleurotus ostreatus virus 1 (PoV1; AY533038), Rosellinia necatrix partitivirus 1 (RnPV1; AB113347), Cryptosporidium parvum virus 1 (CSpV; U95995), Magnaporthe oryzae partitivirus 2 (MoPV2, KX981863.1), Magnaporthe oryzae partitivirus 1 (MoPV1, APP18151.1), Penicillium stoloniferum virus F (PsV-F, AAU95758.1), Fusarium solani virus 1 (FsV1, NP_624350.1), Discula destructiva virus 1 (DdV1; AF316992), Botryotinia fuckeliana partitivirus 1 (BfPV1; AM491609), Ophiostoma partitivirus 1 (OPV1; AM087202), Gremmeniella abietina RNA virus MS1 (GaRV-MS1; AY089993), Penicillium stoloniferum virus S (PsV-S; AY156521), Aspergillus ochraceous virus (AoV; EU118277), rose cryptic virus 1 (RoCV1; EU413666), pepper cryptic virus 2 (PepCV2; JN117278), pepper cryptic virus 1 (PepCV1; JN117276), and Otarine picobirnavirus (oPBV; JQ776552)
References
Cai G, Krychiw JF, Myers K, Fry WE, Hillman BI (2013) A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb dsRNA genome: The sixth member of a proposed new virus genus. Virology 435:341–349
Cai G, Myers K, Fry WE, Hillman BI (2012) A member of the virus family Narnaviridae from the plant pathogenic oomycete Phytophthora infestans. Arch Virol 157:165–169
Cai G, Myers K, Hillman BI, Fry WE (2009) A novel virus of the late blight pathogen, Phytophthora infestans, with two RNA segments and a supergroup 1 RNA-dependent RNA polymerase. Virology 392:52–61
Gillings MR, Tesoriero LA, Gunn LV (1993) Detection of double-stranded RNA and virus-like particles in Australian isolates of Pythium irregulare. Plant Pathol 42:6–15
Hacker CV, Brasier CM, Buck KW (2005) A double-stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J Gen Virol 86:1561–1570
Heller-Dohmen M, Göpfert JC, Pfannstiel J, Spring O (2011) The nucleotide sequence and genome organization of Plasmopara halstedii virus. Virol J 8:123. https://doi.org/10.1186/1743-422X-8-123
Kobayashi S, Uzuhashi S, Tojo M, Kakishima M (2010) Characterization of Pythium nunn newly recorded in Japan and its antagonistic activity against P. ultimum var. ultimum. J Gen Plant Pathol 76:278–283
Lévesque CA, de Cock AWAM (2004) Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res 108:1363–1383
Liu X, Gorovsky MA (1993) Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res 21:4954–4960
Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513–515
Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Suzuki D, Jiang N (2014) Taxonomic reorganization of family Partitiviridae, and other recent progress in partitivirus research. Virus Res 188:128–141
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, Suzuki N, Xie J, Nibert M (2018) ICTV virus taxonomy profile: Partitiviridae. J Gen Virol 99:17–18
Yokoi T, Yamashita S, Hibi T (2003) The nucleotide sequence and genome organization of Sclerophthora macrospora virus A. Virology 31:394–399
Acknowledgments
TM would like to thank Dr. M.J. Roossinck and her laboratory members at Penn State University, USA for discussions that helped inspire this work. We would like to express sincere thanks to Dr. S. Uzuhashi at the Genetic Resources Center, National Agriculture and Food Research Organization, Japan for providing the Pythium nunn isolate.
Funding
This study was funded by the Sasakawa Scientific Research Grant (Sasai, S., #29-409) from The Japan Science Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shiba, K., Hatta, C., Sasai, S. et al. Genome sequence of a novel partitivirus identified from the oomycete Pythium nunn. Arch Virol 163, 2561–2563 (2018). https://doi.org/10.1007/s00705-018-3880-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3880-0