Abstract
The present work reports the discovery and complete genome sequencing of a virus from symptomless radish seedlings, classifiable as a novel member of the genus Alphapartitivirus, family Partitiviridae. Total RNA extracted from germinating seedlings was sequenced using Illumina technology. Bioinformatic analysis of the RNA-seq data revealed two contigs representing the near full-length genomic sequences of two genomic RNAs representing a new virus. Analysis of the genome sequence (excluding the polyA tail, RNA1: 1976 nt and RNA2: 1751 nt, respectively) showed a genomic organization typical of viruses classed within the Partitiviridae, with each genomic RNA encoding a single open reading frame (ORF). Phylogenetic analysis of the RNA dependent RNA polymerase (RNA1 ORF) and of the capsid protein (RNA2 ORF) clearly showed the new virus can be classified within the genus Alphapartitivirus, but sequence divergence establishes it as a new species, for which the name “Raphanus sativus cryptic virus 4” is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viruses classified within the family Partitiviridae have two essential, double-stranded RNA genome segments and have so far been reported in plants, fungi (both basidiomycetes and ascomycetes) and protozoa [1, 2]. Each genome segment, ca. 1300-2500 nucleotides (nt) in length, encodes a single open reading frame (ORF), with RNA1 encoding the viral RNA-dependent RNA polymerase (RdRp) and RNA2 the capsid protein (CP) [1, 2]. The increased availability of genomic sequences has led to a profound taxonomic reorganization of the family into 5 genera, Alpha-, Beta-, Delta- and Gammapartitivirus and Cryspovirus [1], with plant-associated agents found in the first three genera [1, 3]. These five genera are separated on the basis of phylogenetic analyses of the viral RdRp but also differ in some other properties, including the length of genome segments (and of the encoded proteins) and, possibly, in conserved sequences at the plus strand 5’ termini [1]. In plants, partitiviruses are historically referred to as cryptic viruses [4] and are characterized by a persistent life style [5]. Lacking a movement protein, they are unable to move from cell-to-cell and systemically invade host plants, and rather move vertically through the cell division processes of their host [2, 4]. They are seed-transmitted with a high efficiency and generally are not associated with any symptomatology [4].
Material and methods
Virus material
A new alphapartitivirus was identified in radish (Raphanus sativus, unreleased breeding line F1147). Total nucleic acids were extracted from young seedlings collected 72h after seed imbibition in sterile germination boxes. Approximately 250 seedlings were soaked in 50 ml of RNAlater® (Ambion) and mixed softly with a stomacher for 2 min. The suspension was subsequently filtered on 100 µM and 5 µM membranes and the filtrate was centrifuged (10,000 g, 10 min, 4 °C). Total RNAs were extracted from the pellet using a protocol described by Darsonval et al. [6].
Results
Illumina RNA-Seq (HiSeq3000 platform, 2x150bp paired reads) was performed at the Genome-Transcriptome core facilities (Genotoul GeT-PlaGe; France). After demultiplexing and quality trimming, reads were assembled and the obtained contigs annotated using BLASTn and BLASTx against the GenBank database. Two contigs were identified with high homology to the two genomic RNAs of a group of plant-associated viruses classified within the Alphapartitivirus genus, in particular carrot cryptic virus (CCV) [7], beet cryptic virus 1 (BCV1) [8], Vicia cryptic virus (VCV) [9] and the type member of the genus, white clover cryptic virus 1 (WCCV1) [10]. Further analysis of the sequence data [11] and mapping of the reads from the two obtained contigs showed that they incorporated a high proportion of the sequencing reads (from 5.9 to 14.7% depending on the RNASeq library), providing extremely deep genome coverage (on average, 2500x and 3500x for RNA1 and RNA2, respectively). The 5’ genome ends were determined using total RNAs, extracted from 1-month old plants grown from the same seed batch, as templates for 5’ Rapid Amplification of cDNA Ends (RACE, Takara Bio Europe/Clontech, Saint-Germain-en-Laye, France) while the 3’ genome ends were determined by RT-PCR using a PolyT primer and internal primers designed using the two contig sequences.
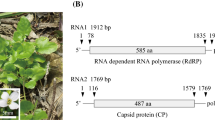
The complete genomic RNAs of the new agent are respectively 1976 nt (RNA1, GenBank MF686921) and 1751 nt (RNA2, GenBank MF686922), sizes typical for viruses classifiable within the Alphapartitivirus [1]. As expected, each genomic RNA encodes a single ORF, corresponding to the RdRp (72.5 kDa; RNA1) and the CP (54.8 kDa; RNA2), respectively (Figure 1), as judged by sequence comparisons with other representative members of the genus. The size of these proteins is also in agreement with expectations for alphapartitiviruses [1] and the 5’ terminal sequence of the two genomic RNAs, 5’GGATCA3’, fits the (n)GAWnW consensus for members of the genus [1]. The 5’ non-coding regions for RNA1 and RNA2 are 93 and 118 nucleotides long, respectively, while the corresponding 3’ non-coding regions are 35 and 163 nucleotides long, respectively. The RdRp contains the expected conserved polymerase motif (cd01699, aa positions 237-439) and shows closest homology (82.9% amino acid (aa) identity, 76.8% nucleotide identity) with the RdRp of CCV and 79.1-82.2% amino acid identity (75.8-76.3% nucleotide identity) with a small group of plant-associated alphapartitiviruses (WCCV1, BCV1, VCV as well as some putative genus members such as dill cryptic virus 1, spinach cryptic virus 1, black grass cryptic virus 1 and red clover cryptic virus 1). By comparison, divergence is much higher with regards to fungus-associated genus members, with only 30.4-58.6% amino acid identity (42.0-62.5% nt identity). A similar range, or even higher, was also observed following comparisons with other partitiviruses, including those previously reported in radishes, such as Rapahanus sativus cryptic viruses 1 [12], 2 [13] and 3 (Genbank FJ461349 and FJ461350) or Raphanus sativus partitivirus 1 (Genbank KT285019) (13.9-32.1% identity). The CP is less conserved, with only 50.6-56.4% aa identity (56.0-61.3% nucleotide identity) with plant-associated alphapartitiviruses (13.5-23.3% amino acid identity and 34.9-41.9% nucleotide identity with fungus-associated genus members. Phylogenetic analyses performed on the RdRp and CP sequences (Figures 2A and 2B) confirmed a clear association with the plant-associated members of the Alphapartitivirus genus, within which the newly identified virus forms a 100% bootstrap-supported subcluster. Interestingly, two expressed sequence tags (EST) from radish (Raphanus sativus var oleiformis, cv. Arena) (EX887526 and EX891659, 701 and 269 nt long, respectively) show 99% nt sequence identity with RNA1 of this virus, indicating that it is present in at least a second radish source, the released Arena variety.
Neighbor-joining phylogenetic trees constructed using the complete amino acid sequences of RNA-dependent RNA polymerases (A) and capsid proteins (B) of various partitiviruses. The trees use strict amino acid identity distances and the statistical significance of branches was evaluated by bootstrap analysis (1,000 replicates). Only values higher than 70% are indicated. The scale bars represents 10% amino acid divergence. Raphanus sativus cryptic virus 4 (RsCV4) proteins are indicated by black dots. The proteins of other viruses classified within the Partitiviridae and associated with radish infections are indicated by a white dot. The following acronyms are used: HPV13: Heterobasidion partitivirus 13; HPV15: Heterobasidion partitivirus 15; HmPVV70: Helicobasidium mompa partitivirus V70; HPV12: Heterobasidion partitivirus 12; HPV3: Heterobasidion partitivirus 3; CpCV1: Chondrostereum purpureum cryptic virus 1; FvBV: Flammulina velutipes browning virus; RsCV1: Raphanus sativus cryptic virus 1; HPV1: Heterobasidion partitivirus 1; CCRSaPV: cherry chlorotic rusty spot-associated partitivirus; BCV1: beet cryptic virus 1; CCV: carrot cryptic virus, WCCV1: white clover cryptic virus 1; VCV: Vicia cryptic virus; RnPV2: Rosellinia necatrix partitivirus 2; RsPV1: Raphanus sativus partitivirus 1; AhV: Atkinsonella hypoxylon virus; WCCV2: white clover cryptic virus 2; CpV1: Cryptosporidium parvum virus 1; PsVF: Penicillium stoloniferum virus F; RsCV2: Raphanus sativus cryptic virus 2; RsCV3: Raphanus sativus cryptic virus 3; SaCV1: Sinapis alba cryptic virus 1; BCV2: beet cryptic virus 2
Taken together, these results show that this virus is clearly distinct from previously characterized partitiviruses and, in particular, from those reported in radish. Our data also indicate that this virus fits all the criteria to be classified as a new species and member of the Alphapartitivirus genus member, with the name “Raphanus sativus cryptic virus 4” proposed. As with other plant-infecting alphapartitiviruses, it is unlikely that Raphanus sativus cryptic virus 4 (RsCV4) has any pathological significance since it was identified in symptomless plants.
References
Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N (2014) Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141
Ghabrial SA, Nibert ML, Maiss E, Lesker T, Baker TS, Tao YJ (2012) Family Partitiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy. 9th Report of the ICTV. Elsevier Academic Press, Amsterdam, pp 523–534
ICTV (2017) ICTV Master species list 2016 v1.3. https://talk.ictvonline.org/files/master-species-lists/m/msl/6776. Accessed 1 Aug 2017
Boccardo G, Lisa V, Luisoni E, Milne RG (1987) Cryptic plant viruses. Adv Virus Res 32:171–214
Roossinck MJ (2010) Lifestyles of plant viruses. Philos Trans R Soc Lond B: Biol Sci 365:1899–1905
Darsonval A, Darrasse A, Durand K, Bureau C, Cesbron S, Jacques M-A (2009) Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans. Mol Plant Microbe Interact 22:747–757
Willenborg J, Menzel W, Vetten HJ, Maiss E (2009) Molecular characterization of two alphacryptovirus dsRNAs isolated from Daucus carota. Arch Virol 154:541–543
Szego A, Enunlu N, Deshmukh SD, Veliceasa D, Hunyadi-Gulyas E, Kuhne T, Ilyes P, Potyondi L, Medzihradszky K, Lukacs N (2010) The genome of Beet cryptic virus 1 shows high homology to certain cryptoviruses present in phylogenetically distant hosts. Virus Genes 40:267–276
Blawid R, Stephan D, Maiss E (2007) Molecular characterization and detection of Vicia cryptic virus in different Vicia faba cultivars. Arch Virol 152:1477–1488
Boccardo G, Milne RG, Luisoni E, Lisa V, Accotto GP (1985) Three seedborne cryptic viruses containing double-stranded RNA isolated from white clover. Virology 147:29–40
Candresse T, Marais A, Faure C, Gentit P (2013) Association of Little cherry virus 1 with the Shirofugen Stunt Disease and genome characterization of a divergent LChV1 isolate. Phytopathology 103:293–298
Chen L, Chen JS, Liu L, Yu X, Yu S, Fu TZ, Liu WH (2006) Complete nucleotide sequences and genome characterization of double-stranded RNA 1 and RNA 2 in the Raphanus sativus-root cv. Yipinghong. Arch Virol 151:849–859
Chen L, Chen JS, Zhang H, Chen SN (2006) Complete nucleotide sequences of three dsRNA segments from Raphanus sativus-root cv. Yidianhong with leaf yellow edge symptoms. Arch Virol 151:2077–2083
Acknowledgements
This work was supported in part by the MetaT Project of the Meta-omics and microbial ecosystems (MEM) INRA Metaprogram. The authors want to thank Catherine Zanchetta and Olivier Bouchez from the GeT core facility (Toulouse, France, http://get.genotoul.fr) for their help in the sequencing experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors (LS, ST, MB and TC) declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ioannis E. Tzanetakis.
The nucleotide sequence reported in this work has been deposited in the GenBank database under accession numbers MF686921 and MF686922.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Svanella-Dumas, L., Theil, S., Barret, M. et al. Complete genomic sequence of Raphanus sativus cryptic virus 4 (RsCV4), a novel alphapartitivirus from radish. Arch Virol 163, 1097–1100 (2018). https://doi.org/10.1007/s00705-017-3693-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3693-6