Abstract
Pepper chlorotic spot virus (PCSV), newly found in Taiwan, was identified as a new tospovirus based on the molecular characterization of its S RNA. In this study, the complete M and L RNA sequences of PCSV were determined. The M RNA has 4795 nucleotides (nts), encoding the NSm protein of 311 aa (34.5 kDa) in the viral (v) strand and the glycoprotein precursor (Gn/Gc) of 1122 aa (127.6 kDa) in the viral complementary (vc) strand. The L RNA has 8859 nts, encoding the RNA-dependent RNA polymerase (RdRp) of 2873 aa (330.8 kDa) in the vc strand. Analyses of the NSm, Gn/Gc and RdRp of PCSV revealed that PCSV is phylogenetically clustered within the watermelon silver mottle virus-related clade. Based on the whole genome sequence, PCSV is closely related to Tomato necrotic ringspot virus and should be classified as a new tospovirus species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Viruses classified within the plant-infecting genus Tospovirus of the family Bunyaviridae are characterized by quasi-spherical enveloped particles of 80-120 nm in diameter. They possess a tripartite genome of three single-stranded RNA segments named L, M and S according to their size [1]. The L RNA is of negative sense and contains a large open reading frame (ORF) encoding an RNA-dependent RNA polymerase (RdRp) in the viral complementary (vc) strand [2]. Both M RNA and S RNA are ambisense, consisting of two ORFs flanked by an A-U rich intergenic region (IGR) that forms a hairpin structure and serves as the transcriptional terminator [3]. The M RNA encodes a movement protein NSm in the viral (v) strand and the glycoproteins Gn and Gc in the vc strand [4, 5]. The v strand of S RNA encodes the gene silencing suppressor NSs [6, 7], and its vc strand encodes the structural nucleocapsid protein (NP) [8]. The classification of tospoviruses is based on their vector specificity, plant host range, serological relationship and the amino acid (aa) identity of the NP, set at a threshold cut-off of lower than 90% [1].
In 2009, a disease causing mottling and deformation on leaves and fruits of sweet pepper was observed in Sinyi Township, Nantou County of central Taiwan. The crude sap of symptomatic sweet pepper samples reacted with antisera against two tospoviruses, capsicum chlorosis virus (CaCV) and watermelon silver mottle virus (WSMoV). A virus isolate, denoted TwPep3, was obtained from the diseased pepper sample and identified as a new tospovirus species belonging to the WSMoV serogroup, provisionally named Pepper chlorotic spot virus (PCSV), according to sequencing of the S RNA and the serological relationship of NP [9]. In this study, the complete sequences of the M and L RNAs of PCSV-TwPep3 were determined and their molecular characteristics were analyzed.
PCSV-TwPep3 was maintained in Chenopodium quinoa Willd. by mechanical inoculation, as described previously [9]. Total RNA from PCSV-TwPep3-infected C. quinoa leaf was isolated using the previously reported method [10]. Degenerate primers were designed based on multiple alignments of available genomic sequences of tospoviruses and specific primers were designed from the determined viral sequences. The accession numbers of the analyzed sequences from GenBank are listed in Supplementary Table 1. The sequences of the primers are shown in Supplementary Table 2. The strategy for amplification of the genomic fragments of the M and L RNAs from PCSV-TwPep3 is illustrated in Supplementary Fig. 1. First strand cDNAs were synthesized from 2 μg of total RNA using SuperScriptTM III RNase H- reverse transcriptase (Invitrogen, CA, USA) and reverse primers (Supplementary Fig. 1; Supplementary Table 2) at 50°C for 60 min, and the reaction was terminated at 75°C for 15 min. PCR amplification of cDNA was done using 2.5 U of TaKaRa LA Taq DNA polymerase (Takara, Kyoto, Japan) with the supplied LA PCR buffer II, 2.5 mM MgCl2, 0.4 mM dNTPs, 200 ng of forward and reverse primers (Supplementary Fig. 1; Supplementary Table 2). PCR amplification was carried out using a thermal cycler (PTC-200, MJ Research Inc., USA), with the following temperature cycle conditions: initial denaturation of 94°C for 5 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min, and one final step at 72°C for 10 min. Viral double-stranded RNAs (dsRNAs) were also purified from virus-infected C. quinoa leaf tissues following the method described previously [11] for verifying the 5′- and 3′-terminal sequences of the M and L RNAs. The dsRNAs were polyadenylated using yeast poly(A) polymerase (Amersham Pharmacia Biotech Inc., NJ, USA) according to the manufacturer’s instruction. These were then used as the templates for RT-PCR amplification of the 5′ and 3′ terminal sequences with an oligo d(T) primer and specific primers designed against previously identified sequences. All amplicons were cloned using the TOPO-pCRII TA cloning kit (Invitrogen) according to the manufacturer’s instructions. Three clones were selected for each fragment and were sequenced by the automatic DNA sequencing method performed at the Biotechnology Centre, National Chung Hsing University, Taichung, Taiwan using an ABI PRISM 3730 DNA sequencer (Applied Biosystems, Hammonton, NJ, USA).
The Lasergene 7 software package (DNASTAR, Madison, WI, USA) was used to analyze sequences. The nucleotide sequences of the M and L RNAs of PCSV-TwPep3 were assembled and analyzed using the SeqMan program. The SeqBuilder program was used to deduce the aa sequence of viral proteins. The GeneQuest and Protean programs were used to analyze the composition and property of nucleic acid bases and proteins. The sequence comparisons were performed using the MegAlign program. For calculating the distance matrices of the viral protein sequences and analyzing phylogenetic relationships, the method of neighbor-joining [12] with 1,000 bootstrap iterations in MEGA 6.0 (Molecular Evolutionary Genetics Analysis software, version 6.0) [13] was used. Tospoviral genomic sequences for comparison with PCSV-TwPep3 were obtained from GenBank (Supplementary Table 1). The SignalP 4.1 server [14], NetNGlyc 1.0 Server [15], NetOGlyc 4.0 Server [16] and TMAP program in SDSC Biology Workbench v3.2 website were used to predict the possible cleavage sites, N-glycosylation sites, O-glycosylation sites and transmembrane domains of the Gn/Gc glycoprotein precursor, respectively. The PROSITE website [17] was used to find functional domains within the L protein.
The results showed that the M RNA (acc. no.: KY315810) was 4795 nucleotides (nts) in length consisting of two ORFs with an ambisense coding strategy. The 5′- and 3′-UTRs of the M RNA are 60 nts and 48 nts in length, respectively. The IGR of M RNA has 382 nts. The NSm coding sequence of PCSV-TwPep3 contains 936 nts, starting from nt 61 to nt 996 in the v strand of the M RNA, encoding a protein of 311 aa (34.5 kDa). Another ORF in the vc strand was 3396 nts long, located between nt 1379 and nt 4747 and encoding the Gn/Gc glycoprotein precursor of 1122 aa (127.6 kDa). The L RNA (acc. no.: KY315809) was determined as being 8859 nts in length and contained an 8622-nt ORF in the vc strand of between nt 30 and nt 8651. The ORF encoded an RdRp of 2873 aa with a predicted molecular mass of 330.8 kDa. Conserved RdRp motifs were found between aa position 1349 and 1529 in the PCSV-TwPep3 RdRp, including motif A (DxxKW, aa 1384-1388), motif B (QGxxxxxSS, aa 1472-1480), motif C (SDD, aa 1510-1512), motif D (K, aa 1557), motif E (ExxS, aa 1567-1570) and motif F (KxQxxxxxR, aa 1303-1311) (Fig. 1). The 5′- and 3′-UTRs of the vc strand of PCSV-TwPep3 L RNA contain 29 nts and 208 nts, respectively.
Multiple alignments of the RNA-dependent RNA polymerase (RdRp) catalytic domains of tospoviruses. Six conserved motifs are shown. Dots and dashes indicate conserved amino acids and gaps within these tospovirus RdRps, respectively. The Bunyaviridae RdRp consensus sequences are represented in bold. The abbreviations of these tospoviruses and the sequences used for this analysis are listed in Supplementary Table 2
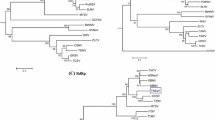
The profiles of the M and L RNAs of PCSV-TwPep3 were compared with those of other tospoviruses, as shown in Supplementary Table 3 and Table 4, respectively. The NSm and Gn/Gc proteins of PCSV-TwPep3 exhibited the highest aa identity of 86.5% and 82.2%, respectively, with proteins encoded by tomato necrotic ringspot virus (TNRV). The PCSV-TwPep3 RdRp shared 72.3-73.9% aa identities with RdRps encoded by WSMoV-serogroup tospoviruses including CaCV, calla lily chlorotic spot virus (CCSV), groundnut bud necrosis virus (GBNV), melon yellow spot virus (MYSV), tomato necrotic spot associated virus (TNSaV), tomato zonate spot virus (TZSV), watermelon bud necrosis virus (WBNV) and WSMoV. Phylogenetic analyses indicated that PCSV is closely related to members of the WSMoV serogroup (Fig. 2).
Phylogenetic analyses of the deduced virus-encoded proteins of Pepper chlorotic spot virus (PCSV) and other tospoviruses. The nonstructural NSm (a), Gn/Gc glycoprotein precursor (b) and RNA-dependent RNA polymerase (RdRp) (c) proteins were analyzed individually. Tospoviruses sharing serological relationships and their serogroups are indicated. The sequences of PCSV analysed in this study are indicated in the boxes. Phylogenetic analyses were done using the neighbor-joining algorithm with 1,000 bootstrap replicates in the MEGA6.0 software. Bootstrap consensus trees are shown. The abbreviations of these tospoviruses and their respective accession codes are listed in Supplementary Table 2
The topology of the Gn/Gc precursors of PCSV-TwPep3 and other tospoviruses was analyzed and compared (Supplementary Fig. 2). In the PCSV-TwPep3 Gn/Gc precursor, two deduced cleavage sites were found at aa 26 (LLS-RT) and aa 436 (SLA-SS). The predicted molecular masses of the mature Gn (aa 27-436, ~45 kDa) and Gc (aa 437-1122, ~75 kDa) were also obtained. Three N-glycosylation sites (two in Gn and one in Gc) and two O-glycosylation sites (one in Gn and one in Gc) were predicted. A total of five transmembrane domains were found, one in the deduced signal peptide, three in the mature Gn portion, and one in the C-terminal region of the Gc protein. Tospoviruses grouped in the same serogroup normally share conserved cleavage sites within their Gn/Gc precursors. The first deduced cleavage sites of the Gn/Gc precursors of PCSV-TwPep3 (LLS-RT) and TNRV (SFA-SE) are distinct from the conserved cleavage sequence “V(Y/F)L-L(N/S/K)” found in other members of the WSMoV serogroup. Of note, the sequence “VYLLS” neary the first predicted cleavage site in the Gn/Gc precursors of PCSV-TwPep3 and TNRV could represent a cleavage site.
The whole genome sequence of PCSV-TwPep3 has been completed. According to molecular analysis of the genome and phylogenetic analysis of individual viral proteins, PCSV-TwPep3 is a tospovirus species closely related to TNRV.
References
King AMQ, Lefkowitz E, Adams MJ, Carstens EB (2012) Virus taxonomy: 9th report of the international committee on taxonomy of viruses. Elsevier, New York
de Haan P, Kormelink R, de Oliveira Resende R, van Poelwijk F, Peters D, Goldbach R (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol 72:2207–2216
Pappu SS, Bhat AI, Pappu HR, Deom CM, Culbreath AK (2000) Phylogenetic studies of tospoviruses (Family: Bunyaviridae) based on intergenic region sequences of small and medium genomic RNAs. Arch Virol 145:1035–1045
Kormelink R, Storms M, Van Lent J, Peters D, Goldbach R (1994) Expression and subcellular location of the NSM protein of tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200:56–65
Storms MMH, Kormelink R, Peters D, Van Lent JWM, Goldbach RW (1995) The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 214:485–493
Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett 532:75–79
Bucher E, Sijen T, de Haan P, Goldbach R, Prins M (2003) Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J Virol 77:1329–1336
de Haan P, Wagemakers L, Peters D, Goldbach R (1990) The S RNA segment of tomato spotted wilt virus has an ambisense character. J Gen Virol 71:1001–1007
Cheng YH, Zheng YX, Tai CH, Yen JH, Chen YK, Jan FJ (2014) Identification, characterisation and detection of a new tospovirus on sweet pepper. Ann Appl Biol 164:107–115
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289
Valverde RA, Dodds JA, Heick JA (1986) Double-stranded ribonucleic acid from plants infected with viruses having elongated particles and undivided genomes. Phytopathology 76:459–465
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Gupta R, Jung E, Brunak S (2004) Prediction of N-glycosylation sites in human proteins. http://www.cbs.dtu.dk/services/NetNGlyc/
Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Paul Bennett E, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H (2013) Precision mapping of the human O-GalNAc glycoproteome through simple cell technology. EMBO J 32:1478–1488
Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA (2006) The PROSITE database. Nucleic Acids Res 34:D227–D230
Acknowledgements
We are grateful to Dr. Chung-Jan Chang, Professor Emeritus of the University of Georgia and Dr. Kuang-Ren Chung for their critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
I have read and have abided by the statement of ethical standards for manuscripts submitted to Archives of Virology.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This study was partially supported by a grant (NSC 102-2628-B-005-005-MY3) from the National Science Council of Taiwan and a grant (MOST 105-2313-B-005-021-MY3) from the Ministry of Science and Technology, Executive Yuan, Taiwan.
Additional information
K.-S. Huang and C.-H. Tai contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, KS., Tai, CH., Cheng, YH. et al. Complete nucleotide sequences of M and L RNAs from a new pepper-infecting tospovirus, Pepper chlorotic spot virus. Arch Virol 162, 2109–2113 (2017). https://doi.org/10.1007/s00705-017-3260-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3260-1