Abstract
In this report, we present the first description of the complete genome sequence of a new monopartite begomovirus isolated from tomatoes collected in Burkina Faso and presenting with symptoms of tomato leaf curl disease. We propose the tentative name “tomato leaf curl Burkina Faso virus’’ (ToLCBFV). DNA-A-like nucleotide sequence of ToLCBFV shares the highest nucleotide sequence identity (85%) with the pepper yellow vein Mali virus (PepYVMLV). Phylogenetic analysis confirmed the affiliation of ToLCBFV to Old World monopartite begomoviruses. This discovery of a new species confirms the existence of high genetic diversity in monopartite begomoviruses in sub-Saharan Africa and particularly in West Africa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Annotated sequence record
Begomoviruses (genus Begomovirus, family Geminiviridae) are a group of plant viruses that are transmitted by the whitefly Bemisia tabaci (family Aleyrodidae) and are responsible for serious diseases in a wide range of cultivated crops, including tomato (Solanum lycopersicum). These viruses have a circular single stranded DNA genome encapsidated in twinned icosahedral particles [9].
Since the late 1980s, begomoviruses have been responsible for many emerging vegetable and food crop diseases worldwide [6]. Except for a small number of bipartite begomoviruses described in the Old World, African begomoviruses are monopartite with a unique DNA-A-like component [3]. Today, a complex of more than ten monopartite begomoviruses has been described in tomato in Africa (for reviews see [4, 7]).
In March 2013, leaf samples from three tomato plants showing severe symptoms of leaf curling, resembling tomato leaf curl disease (ToLCD; Figure 1), were collected at Loumbila (latitude: 12.490932; longitude: −1.398806; Figure 1), located 25 km from Ouagadougou, in Burkina Faso. Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). Viral genomes were amplified by rolling-circle amplification using Phi29 DNA polymerase. Amplified products were digested with BamHI endonuclease. The monomeric full-length DNA molecules obtained (~3 kb) were purified and ligated into the pGEM-3Zf vector (Promega). The ligated products were then cloned into Escherichia coli (JM109, Promega). Selected clones were completely sequenced by primer walking (Macrogen) and contigs were assembled with DNA Baser v.2.91 (Heracle BioSoft). Nucleotide sequences were subjected to a BLAST search for preliminary species assignment. Multiple sequences were aligned using the MUSCLE alignment method and maximum-likelihood (ML) phylogenetic trees were constructed by using FastTree v2.1.7. Pairwise identity comparisons of nucleotide sequences were performed using SDT v1.2 with pairwise deletion of gaps [5].
Three DNA-A-like sequences of 2,784 nucleotides in length and 100% identical to each other, were obtained from the same tomato sample. No DNA-B component or betasatellite DNA was detected by PCR with universal primers ([2, 8]). The DNA-A-like component presented features typical of Old World begomoviruses, with six open reading frames (ORFs): V1 (258 aa; capsid protein [CP]), V2 (116 aa; movement protein [MP]) in the virion sense, and C1 (359 aa; replication-associated protein [REP]), C2 (135 aa; transcriptional activator protein [TrAP]), C3 (134 aa; replication enhancer protein [REn]) and C4 (96 aa; C4 protein) in the complementary sense (Supplementary table 1). The DNA-A-like component presented a stem-loop structure in the origin of replication with the canonical nonanucleotide motif (5’-TAATATTAC-3’), TATA and GC boxes and two GGGGT iteron sequences (Supplementary figure 1). The corresponding iteron-related domain (IRD; Rep N-terminal domain interaction with ori-associated iterons) was identified as MAPPKRFRVN [1].
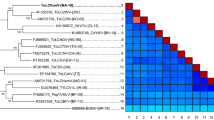
The three DNA-A-like sequences shared the highest nucleotide sequence identity (85%) with a pepper yellow vein Mali virus isolated from Capsicum frutescens in Burkina Faso (PepYVMV-[BF:Ban:Hpe:09], FN555174; supplementary tables 2 and 3; [10]). In accordance with the recently updated begomovirus species demarcation criteria (91% nucleotide identity of DNA-A and DNA-A-like without gaps; [3]), this isolate should be assigned to a new species of tomato-infecting monopartite begomovirus. ML tree analysis confirmed that the complete nucleotide sequence of ToLCBFV forms a distinct branch together with PepYVMLV among current African begomoviruses (Figure 2). In the absence of symptoms that differ from the other African tomato-infecting begomoviruses, we propose the name ‘‘tomato leaf curl Burkina Faso virus’’ (ToLCBFV).
Maximum-likelihood phylogenetic tree and colour-coded matrix of pairwise nucleotide identity inferred from alignments of complete DNA-A-like sequences of ToLCBFV and selected Old World monopartite begomoviruses originating from Africa. Only bootstrap values higher than 70% are shown (1000 replicates). The matrix uses a discontinuous range of three shades of colour (red, green and blue) differentiating two cut-off values representing the strain (93-94%, green-red) and the species (90-91%, blue-green) demarcation thresholds of begomoviruses. For begomovirus acronyms, see supplementary table 3
Taken together, our results demonstrate that ToLCBFV is representative of a new species, genetically distinct from the currently described West African tomato-infecting monopartite begomoviruses, such as ToLCGV [4]. The infectivity and biological features of this new species should be investigated in the future.
GenBank accession number
KX853168
References
Arguello-Astorga GR, Ruiz-Medrano R (2001) An iteron-related domain is associated to Motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid–base pairs by a comparative approach. Arch Virol 146:1465–1485
Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG (2002) Universal primers for the PCR-mediated amplification of DNA beta—a molecule associated with some monopartite begomoviruses. Mol Biotechnol 20:315–318
Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JCF, Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A (2015) Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160:1593–1619
Leke WN, Mignouna DB, Brown JK, Kvarnheden A (2015) Begomovirus disease complex: emerging threat to vegetable production systems of West and Central Africa. Agric Food Secur 4:1–4
Muhire BM, Varsani A, Martin DP (2014) SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. Plos One 9:1–8
Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S (2011) Emerging virus diseases transmitted by whiteflies. Ann Rev Phytopathol 49:219–248
Rey MEC, Ndunguru J, Berrie LC, Paximadis M, Berry S, Cossa N, Nuaila VN, Mabasa KG, Abraham N, Rybicki EP, Martin D, Pietersen G, Esterhuizen LL (2012) Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in Southern Africa, including the South-West Indian Ocean Islands. Viruses 4:1753–1791
Rojas MR, Gilbertson RL, Russel DR, Maxwell DP (1993) Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted geminivirus. Plant Dis 77:340–347
Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43:361–394
Tiendrebeogo F, Lefeuvre P, Hoareau M, Traore VS, Barro N, Perefarres F, Reynaud B, Traore AS, Konate G, Lett JM, Traore O (2011) Molecular and biological characterization of pepper yellow vein Mali virus (PepYVMV) isolates associated with pepper yellow vein disease in Burkina Faso. Arch Virol 156:483–487
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the EMEB project (PEERS-AIRD, 2013-2014), the European Union (FEDER), the Région Réunion and CIRAD. AO is a recipient of a PhD fellowship from CIRAD (2015-2017).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2017_3231_MOESM2_ESM.xls
Supplementary table 2. Percentage of pairwise nucleotide identities shared between full DNA-A-like sequences of ToLCBFV and selected Old World monopartite begomoviruses originating from Africa and in the case of ToLCV, from Australia. Percentages were determined using SDT v1.2, calculated as 1-p distance, with pairwise deletion of gaps [5]. Abbreviations and accession numbers of viruses are listed in supplementary table 3 (XLS 29 kb)
705_2017_3231_MOESM3_ESM.xls
Supplementary table 3. Name, acronyms and GenBank/EMBL/DDBJ accession numbers of begomoviruses used in pairwise identity comparisons and phylogenetic analysis (XLS 32 kb)
Rights and permissions
About this article
Cite this article
Ouattara, A., Tiendrébéogo, F., Lefeuvre, P. et al. Tomato leaf curl Burkina Faso virus: a novel tomato-infecting monopartite begomovirus from Burkina Faso. Arch Virol 162, 1427–1429 (2017). https://doi.org/10.1007/s00705-017-3231-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3231-6