Abstract
Polydnaviruses (PDVs) play a critical role in altering host gene expression to induce immunosuppression. However, it remains largely unclear how PDV genes affect host genes. Here, the complete genome sequence of Microplitis bicoloratus bracovirus (MbBV), which is known to be an apoptosis inducer, was determined. The MbBV genome consisted of 17 putative double-stranded DNA circles and 179 fragments with a total size of 336,336 bp and contained 116 open reading frames (ORFs). Based on conserved domains, nine gene families were identified, of which the IκB-like viral ankyrin (vank) family included 28 members and was one of the largest families. Among the 116 ORFs, 13 MbBV genes were expressed in hemocytes undergoing MbBV-induced apoptosis and further analyzed. Three vank genes (vank86, vank92, vank101) were expressed in hemocytes collected from Spodoptera litura larvae parasitized by M. bicoloratus, in which host NF-κB/IκBs, including relish, dorsal, and cactus, were also persistently expressed. When Spli221 cells were infected with MbBV viral particles, mRNA levels of host and viral NF-κB/IκB genes were persistent and also varied in Spli221 cells undergoing virus-induced pre-apoptosis cell from 1 to 5 hours postinfection. Both were then expressed in a time-dependent expression in virus-induced apoptotic cells. These data show that viral IκB-like transcription does not inhibit host NF-κB/IκB expression, suggesting that transcription of these genes might be regulated by different mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Delivery of viral particles and eggs to the host hemocoel involves the development of immature parasitoid offspring through the stages of eggs, embryos, first-instar, second-instar, and third-instar larvae, which must occur in the absence of the ability of the larva to protect itself against the host cellular immune response. Braconid and ichneumonid wasps that carry polydnaviruses (PDVs) [37] require the immunosuppressive function of a virus to avoid host non-self recognition. The viral family Polydnaviridae is divided into two genera, Bracovirus and Ichnovirus. Bracoviruses (BVs) and ichnoviruses (IVs) have independent origins, but share a similar life style, probably through a convergent evolutionary process [40]. The proviral form of the viral genome is transmitted vertically in wasps; viral DNA replication only occurs at the pupal-adult stage in ovarian calyx cells in the female wasp [1]. Mature virus particles are stored in the lumen of the calyx and oviduct and are injected along with the egg into the lepidopteran host hemocoel. The viral genome is then integrated into the wasp’s host chromosome(s) to transcribe the viral genes to inhibit host immune responses to ensuring proper development of the wasp larva [3, 6, 12, 35–37]. These immunosuppressive processes are controlled mainly by viral genome transcription and its effect on the expression of host genes.
Several BV genomes have been sequenced. The viral genome sizes range from 200 kilobase pairs (kbp) in Microplitis demolitor bracovirus (MdBV) to 600 kbp in Cotesia vestalis bracovirus (CvBV) in separate circles: 15 circles in MdBV and 35 circles in CvBV. The number of predicted open reading frames (ORFs) varies from 61 in MdBV to 157 in CvBV [8, 40]. Over 20 protein families have been identified from BVs, Cotesia congregata bracovirus (CcBV) (total 80 genes in 16 protein families), CvBV (total 91 genes in 17 protein families), Glyptapanteles indiensis bracovirus (GiBV) (total 87 genes in 16 protein families), G. flavicoxis bracovirus (GfBV) (total 79 genes in 15 protein families), and MdBV (total 37 genes in 7 protein families) [10, 11, 14]. However, it is not known exactly which functional genes are expressed in virus-infected host hemocytes.
Of the few protein families common to both BV and IV, viral ankyrins (vanks), occur in members of both genera [40]. Various reports have shown that vanks function as IκB mimics, but they lack a regulatory domain for phosphorylation and ubiquitination, which could result in irreversible binding to NF-κB to inhibit an immune response [4, 7, 38, 39]. IκB-like proteins, containing ankyrin-repeat domains (Anks) that are highly similar to Drosophila IκB, and Cactus, have also been found in BVs, such as CcBV (8 vanks), CvBV (8 vanks), GiBV (9 vanks), GfBV (8 vanks), and MdBV (12 vanks). A recent study demonstrated that two IκB mimics, Ank-H4, and N5, from MdBV bind to Drosophila NF-κB factors Dorsal and Relish, whereupon the expression of several antimicrobial peptide genes is reduced [7].
NF-κB, Relish, and Dorsal share an REL homology domain (RHD), which is essential for dimerization and DNA binding [7, 16, 37]. Human NF-κB/IκB proteins contain two-antiparallel α-helices and a β-hairpin, sharing G-TPLH and LL–GA consensus repeats [28]. Although vankyrins are known to inhibit host NF-κB, their effect on host NF-κB gene expression during viral immunosuppression remains largely unclear. Induction of apoptosis is an effective immunosuppression strategy, and NF-κB/IκB signaling pathways are known to regulate cell survival and apoptosis. Thus, the transcription patterns of NF-κB/IκB in the apoptotic cells can be used to identify vankyrins that interfere with host NF-κB/IκB gene expression.
The Microplitis bicoloratus and Spodoptera litura model is an excellent system for studying immunosuppression occurring via the induction of apoptosis [25–27]. In our previously study, we showed, using gel electrophoresis, that Microplitis bicoloratus bracovirus (MbBV) contains at least 11 circular dsDNAs from 8 to 50 kbp [26]. MbBV-induced apoptosis against host granulocytes causes immunosuppression [25] via viral gene expression in host hemocytes [20]. Transcriptomes of non-parasitized and parasitized S. litura hemocytes contains sequences of host NF-κB/IκB genes, relish, dorsal, and cactus, and also the transcribed viral genes [20]. Importantly, the endogenous Spli221 cell line, which undergoes low-level apoptosis under normal cell culture conditions [21], allows us to generate pre-apoptotic Spli221 cells infected by MbBV particles to perform in vitro assays. To screen for genes expressed in apoptotic hemocytes, we performed whole-genome sequencing of MbBV and identified 116 genes. By comparing those sequences with the transcriptome of S. litura hemocytes parasitized by the wasp M. bicoloratus, we identified three IκB-like genes, vank86, vank92, and vank101, in 13 screened expressed genes. Then, we used a bioinformatics approach to identify NF-κB/IκB-related factors from both the host S. litura and BVs. Using in vivo and in vitro transcriptional analyses of these NF-κB/IκB factors, we identified potential correlations in their expression patterns. We found that viral IκB-like transcription could not inhibit host NF-κB/IκB expression at the mRNA level, suggesting that they might use different mechanisms to regulate transcription.
Materials and methods
Insect rearing and experimental animals
The S. litura colony was reared on an artificial diet formulated as described previously [19] at 27 ± 1 °C, RH 60-80 %, with a 12:12 h photoperiod regime. The parasitoid M. bicoloratus colony was maintained on S. litura larvae reared in the laboratory. Adults were also provided with honey as a dietary supplement. The parasitoid colony was passaged according to established methods [27].
Cell culture
Spli221 (TUAT-Spli221) adherent cells were derived from S. litura [29]. Cells were cultured in TNM-FH insect culture medium containing 10 % fetal bovine serum (FBS, Hyclone). Cells were maintained at 27 °C and passaged in 25-cm2 tissue culture flasks (Corning).
Isolation and purification of viral particles and infection of Spli221 cells
Purified viral particles were prepared based on a previously published protocol [25, 26] with minor modifications. Briefly, fresh wasps were frozen at -20 °C for 10 min and then put on ice. Reproductive tracts of female wasps were excised under a binocular stereo dissecting microscope, and separated ovaries were collected into a 2-ml tube on ice until further use. The calyces were punctured using forceps, and the calyx fluid was resuspended in 1x PBS, and then using a 2.5-ml needle, dispersed by aspirating back and forth. The mixture was centrifuged for 3 min at 1,000 g at 4 °C, to remove eggs and cellular debris. A 0.45-μm syringe filter was used to purify the viral particles. Two hundred thousand Spli221 cells were seeded in a 12-well culture plate (Corning) 2 h before infection. The quantity of virus used in experiments is expressed in wasp equivalents. A volume of 15 µl of purified viral particles (1.5 wasp equivalents) was added per well.
Isolation, sequencing, assembly and gene prediction of viral genomic DNA
Purified viral supernatant was centrifuged at 12,000 g for 15 min at 4 °C, and viral pellets were incubated in 200 µl of PDV-DNA lysis buffer (100 mM NaCl, 10 mM Tris/HCl, 25 mM EDTA, 0.5 % SDS, pH 8.0) containing 2.5 mg of proteinase K per ml, 8 µl of 20 % Sarcosyl solution, and 1 mg of RNaseA per ml at 55 °C for 3 h. The isolated DNA was further purified by phenol-chloroform extraction and subsequent ethanol precipitation. High-quality DNA samples (with an A260/A280 ratio>2.0) were further amplified using an illustra™ TempliPhi kit (GE Healthcare UK) following the manufacturer’s instructions. The samples were sequenced using an Illumina Hiseq 2000, and the total number of bases sequenced was greater than 3 Gbp. De novo DNA-seq assembly was performed using Velvet and ABySS software, respectively [34, 42]. Assembled contigs were merged, and redundant sequences were removed. GeneMark software was used to identify functional proteins from the isolated contigs [24].
PDV gene expression in hemocytes
Clean reads were mapped to assembled contigs to obtain RPM values based on read numbers [30]. Statistical analysis of data was performed using DESeq [2]. A p-value of 0.001 was set as the criterion for identification of significant differences in gene expression based on a previously described method [20].
Isolation of total RNA, cDNA synthesis, and qRT-PCR
Whole tiny larvae collected 1–3 days post-parasitization (p.p.), hemocytes from 4-7 days p.p. collected from parasitized S. litura larvae, and Spli221 cells collected 1-5 h postinfection (p.i.) were used for RNA isolation. Isolation of total RNA, cDNA synthesis, and qRT-PCR was performed as per previously published protocols [20]. The 2-∆∆CT method was used as reported previously [22]. Three replications were carried out for each sample. Assays were repeated at least three times. Comparisons were performed using unpaired t-tests.
Phylogenetic and transcriptional analysis of NF-κB and IκB-like genes
The amino acid sequences of NF-κB and IκB-like proteins from S. litura, M. bicoloratus bracovirus, and M. demolitor bracovirus were retrieved from GenBank and aligned using webPRANK [23]. Alignment of NF-κB and IκB-like sequences was performed for phylogenetic analysis using the maximum-likelihood method. These analyses were performed using MEGA 6 software [17]. The transcriptional data were used for further analysis to generate a heatmap using MeV v4.9 software [33].
Statistical analysis
Comparisons between data groups were performed as stated in each figure legend using GraphPad Prism ver. 6. Differences between groups with a p-value less than 0.05 were considered significant. The resulting data are presented as mean ± SEM. All assays were repeated at least three times.
GenBank accession numbers
All of the sequences from this project have been deposited in the GenBank database under the accession numbers KP258410 to KP258647 and KP274920.
Results
General features of the M. bicoloratus bracovirus (MbBV) genome
After sequencing, a total of 7,620,970 paired reads were obtained, with 250-bp-length sequences from each pair and a total sequence length of 3.8 Gbp. Altogether, 336,336 bp of MbBV genomic fragments were obtained from 239 reassembled scaffolds ranging in length from 500 base pairs (bp) to 15,413 bp (GenBank accession nos. KP258410-KP258647 and KP274920). In total, 116 genes were identified (Table 1), and 60 contigs were mapped to the sequenced genomes from three other bracoviruses, namely M. demolitor bracovirus, C. congregata bracovirus, and C. sesamiae bracovirus (CsBV). Computational comparison of the genomic organizations of the three viral genomes suggested that the genes identified in MbBV were organized in 17 putative dsDNA circles (Table 2). Therefore, the putative circles were named MbBV-C1 to C17 (Fig. 1A). The remaining contigs (F1-F179) were fragments that did not form parts of a circle (Table 1, Figs. S1, S2). Computational analysis of the putative circles and fragments revealed the presence of 50 ORFs in 15 circles (Fig. 1A) and 66 ORFs in 61 fragments (Fig. 1B).
The genome of M. bicoloraus bracovirus. The diagram represents the properties of genomic fragments of MbBV. Seventeen potential circular segments containing 50 unigenes (A) and 61 out of a total 179 fragments, containing 66 viral genes, (B) are shown. The numbers in brackets indicate the number of genes, while asterisks indicate viral genes expressed in hemocytes of S. litura parasitized by M. bicoloratus
General features of the predicted ORFs in the MbBV genome
In the MbBV genome, we identified 116 ORFs, which we numbered according to the length of the scaffold on which they were found (Table 1). Every single ORF was predicted to encode a protein of > 100 amino acids in length. In total, 14 of the putative circles contained ORFs, except circles 2 and 4, while circles 7 and 10 contained only one ORF, and circle 16, which was the largest genomic segment, contained 10 ORFs. Fragment 161 contained three ORFs, fragments 141, 142, and 155 contained two ORFs, and 57 fragments each contained only one ORF (Fig. 1B). No ORFs were found in the remaining 118 fragments (Figs. S1, S2). These 116 genes were then analyzed on the basis of the presence of conserved domains. A unique feature of the MbBV genome is the major gene families it encodes. In MbBV, nine gene families were identified. Table 3 provides an overview of the predicted proteins, together with their corresponding protein families. With 28 members, ankyrin-repeat (Vank) and protein tyrosine phosphatases (PTPs) were the two largest protein families we identified. The third-largest MbBV gene family we identified was reverse transcriptase (RT), which comprised five gene members. RT genes are a new gene family found in BVs, and their functions in these viruses are unknown. The fourth-largest MbBV gene family we identified was the Ben-domain-coding proteins, which comprise three gene members. Aminoglycoside phosphotransferase (APH), and N-methylhydantoinase (ATP hydrolyzing) are two new families found in BVs, and their functions are also unknown. The remaining three gene families, EGF-like, mucin-like (a Glc) and helicase each comprise one member. The 47 hypothetical protein members await further analysis. The genes were named by using the protein family plus gene ID, as shown in Table 4, which contains the gene ontology (GO) annotation required for further functional research. Finally, the 28 viral ankyrin genes, which are commonly shared by BVs and IVs, were similar to the IκB-like gene.
Comparative transcriptome analysis of host NF-κB/IκB and MbBV genes transcribed in apoptotic hemocytes by parasitism
The transcriptome data for hemocytes undergoing apoptosis after parasitization by M. bicoloratus and non-apoptotic hemocytes have been reported previously [20]. To screen for the viral genes transcribed in host hemocytes that are involved in apoptosis, we compared the 116 genes identified in MbBV with the transcriptome of host-parasitized apoptotic hemocytes. The two generated datasets revealed that 13 viral genes were expressed in the apoptotic hemocytes upon parasitism by the wasp M. bicoloratus (Table 5). These genes, which belong to six conserved gene families in the MbBV genome, include three vank genes, six ptp genes, one hp gene, one ben gene, one egf-like gene, and one mucin-like gene (Fig. 1A and B, asterisks). The three proteins that contained ankyrin-repeat domains were Vank86 in circle 14, Vank92 in circle 10, and Vank101 in circle 16 (Fig. 1A). RNAseq-based comparative analysis and hierarchical cluster analysis revealed that the abundance of mRNA for viral genes such as IκB-like, Vank86, Vank92 and Vank101 were enriched in the apoptotic hemocyte transcriptomes of parasitized S. litura (RPKM_M), but not in host NF-κB/IκB, Relish, Dorsal, and Cactus (Fig. 2, red asterisks).
Comparative transcriptome analysis of the viral IκB-like genes and host NF-κB/IκB hemocytes in which apoptosis was induced by natural parasitism. Hierarchical cluster analysis shows NF-κB/IκB and viral IκB-like genes transcribed in the hemocytes of S. litura (RPKM_S) and hemocytes parasitized by M. bicoloratus (RPKM_M). RPKM: Reads per kilobase of exon model per million mapped reads. Red asterisks show the host NF-κB, Relish, Dorsal, and Cactus and viral IκB-like, Vank86, Vank92, and Vank101
Functional domain analysis of viral IκB-like and host NF-κB/IκB genes transcribed in apoptosis-induced hemocytes
To obtain a detailed breakdown of the Ank domains present in both virus and host, we performed a bioinformatics analysis of the different domains based on their ORF sequences. For the functional analysis, we also analyzed two Ank proteins from MdBV, namely N5 and H4, whose IκB-like functions have been studied before [38]. The phylogenetic tree obtained from the analysis followed the alignment of the two MdBV genes and the three Ank domain genes from MbBV, as well as the three host NF-κB/IκB genes, dorsal, relish, and cactus. As shown in Fig 3A, the phylogenetic tree was separated into two branches, one containing the two NF-κB host gene members, and one containing all of the other genes that are related to IκB (Fig. 3A, left), and the gene domains based on the ORFs are also shown (Fig. 3A, right). Host IκB Cactus contained 10 Ank domains: three Ank, three Ank2, and four Ank4. Vank92 and H4 contain two Ank domains. Vank101, N5, and Vank86 contain four Ank domains. Host Relish contains three Ank2 and two Ank domains, the N-terminal subdomain of the RHD, which is involved in DNA binding, and the IPT domain of the transcription factor NF-κB and the death domain of NF-κB precursor proteins. Host Dorsal does not harbor an Ank domain, but it contains an MADF, DNA-binding domain at its N-terminus and a BESS motif at its C-terminus. All Ank domains were separated from host IκB and viral IκB-like and then aligned. For the IκB branch, the Ank superfamily, which contains two antiparallel α-helices and a β-hairpin [28], was aligned and the similar consensus sequence (-LL–GAD/N, -G-TPLH-) identified may be involved in the formation of α-helices (Fig. 2B). A similar analysis of the viral proteins shows that Vank86 contains three Ank domains and one Ank2 domain, and that Vank92 contains two overlapping Ank domains, while Vank101 contains two Ank and two Ank2 domains. However, none of these viral proteins contain an RHD, suggesting that their role might be similar to that of IκB (i.e. an inhibitor of NF-κB function). Two specific domains, IPT_NF-κB in the N-terminus and Death_NF-κB in the C-terminus of Relish, are shown in Fig. 3C. Taken together, these results suggest that Vank86, Vank92, and Vank101 from MbBV and H4, and N5 from MdBV are highly similar in their overall sequences, and importantly, they share the same Ank motifs. This indicates that they are likely to act as viral IκB mimics.
Analysis of functional domains of viral IκB-like genes and host NF-κB/IκB. (A) Phylogenetic tree of the NF-κB/IκB and IκB-like family built using MeV software and Mega6. The maximum-likelihood tree was built on the basis of NF-κB/IκB and IκB-like domains. The amino acids clustering together were divided into two distinct subclasses: NF-κB, Relish, and Dorsal from S. litura; IκB, Cactus from S. litura, IκB-like, Vank86, Vank92 and Vank101 from M. bicoloratus bracovirus; and H4 and N5 from M. demolitor bracovirus. (B) Alignment of Ank domains of NF-κB/IκB and IκB-like proteins. Two antiparallel α-helices similar to the consensus sequences are shown. (C) NF-κB domains in Relish: an IPT_NF-κB domain at the N-terminus and a Death_NF-κB domain at the C-terminus. (D) DNA binding domains in Dorsal: MADF_DNA_bdg at the N-terminus and a BESS motif in the C-terminal
Molecular interactions between host and virus with respect to NF-κB signaling in apoptotic hemocytes
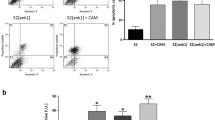
To investigate the expression patterns of both host and viral genes of the NF-κB/IκB family in apoptotic hemocytes, we compared the mRNA expression levels for both viral IκB-like and host NF-κB/IκB genes. Total RNA from larvae at 1-3 days post-parasitization (p.p.), as well as from the apoptotic hemocytes at 4-7 days p.p., was isolated, and qRT-PCR was performed (Table 6, Figs. S3-S9). Fig. 4A shows how host NF-κB Relish increased from 1 to 3 days p.p. However, no significant differences were seen in the mRNA levels among the samples from 4, 5, 6 and 7 days p.p.. By comparison, host Dorsal showed a statistically significant decrease between days 1 and 2 p.p. (Fig. 4B). The Dorsal mRNA level in the apoptosis-induced hemocytes was relatively stable, with no significant differences observed for days 4 to 7 p.p.. For host IκB Cactus, the hemocyte mRNA level increased markedly between days 4 and 7 p.p., and the mRNA levels in the larvae from days 1 to 3 p.p. were consistently stable (Fig. 4C). The viral IκB-like Vank86 had significantly lower mRNA levels in larvae on days 1 and 2 p.p. compared with the levels on days 3 p.p. onwards (Fig. 4D). In contrast, the mRNA expression levels of the viral IκB-like genes Vank92 and Vank101 were maintained at a stable level from days 1 to 3 p.p. (Fig. 4D-F). Three of the IκB-likes (Vank86, Vank92, and Vank101) maintained relatively stable levels from days 4 to 7 p.p. (Fig. 4D-F). Together, these results suggest that viral IκB-like genes and host NF-κB/IκB genes are expressed at the same time, which also suggests that viral IκB-like genes did not inhibit the mRNA level of host NF-κB/IκB during wasp larvae development in the virus-induced apoptotic hemocytes in the host hemocoel.
Expression patterns of viral IκB-like genes and NF-κB/IκB in hemocytes in which apoptosis was induced by natural parasitism. Quantitative real-time PCR analysis was performed from 1 to 7 days post-parasitization (p.p.). Normalized mRNA levels (fold change) of Relish (A), Dorsal (B), Cactus (C), Vank86 (D), Vank92 (E), and Vank101 (F) are shown. Data are shown as mean ± SEM. Comparisons were performed using unpaired t-tests. 1 day p.p. compared with 2 and 3 day p.p. of parasitized larvae, and 4 day p.p. compared with 5 to 7 day p.p. of parasitized hemocytes, respectively. * indicates p < 0.05. ** indicates p < 0.01. *** indicates p < 0.001
Host-virus molecular interactions with respect to NF-κB signaling in Spli221 pre-apoptotic cells
To investigate further the expression patterns in the early infection stage between viral IκB-like and host NF-κB/IκB proteins in a virus-induced apoptotic cell population, we isolated fresh MbBV particles and use them to directly infect Spli221 cells derived from S. litura [41], a process that allowed us to generate a pre-apoptotic cell population with which to perform in vitro assays. Total RNA, isolated from cells at 1-5 h postinfection (p.i.), was subjected to qRT-PCR. The expression pattern of host NF-κB from Relish decreased significantly from 1 h to 2 h p.i. and from 4 h to 5 h p.i. (Fig. 5A). In contrast, host NF-κB mRNA levels from Dorsal (Fig. 5B) and host IκB from Cactus (Fig. 5C) did not differ significantly. Fig. 5D shows that viral IκB-like Vank86 mRNA levels significantly increased between the 1 h and 3 h samples and between 1 h and 4 h samples p.i. Intriguingly, viral IκB-like Vank101 mRNA levels between 1 h and 3 h and between 1 h and 4 h p.i. decreased significantly (Fig. 5E). However, no expression of viral IκB-like Vank92 was observed over the short time period of infection. Importantly, these results suggest that the viral IκB-like genes Vank86 and Vank101 are transcribed with host NF-κB/IκB at the same time soon after infection with MbBV particles. This result implies that viral IκB-like mRNA does not inhibit host NF-κB/IκB mRNA expression and that viral IκB-like genes and host NF-κB/IκB genes may use different transcription mechanisms.
Expression patterns of viral IκB-like genes and NF-κB/IκB Spli221 cells in which pre-apoptosis was induced by infection with MbBV particles. Quantitative real-time PCR analysis was performed from 1 to 5 hours postinfection (p.i.). Normalized mRNA levels (fold change) of Relish (A), Dorsal (B), Cactus (C), Vank86 (D) and Vank101 (E) are shown. Data are represented as mean ± SEM. Comparisons were performed using unpaired t-tests: 1 hour p.i. compared with different time points, respectively.* indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001
Discussion
Polydnavirus transcription is thought to result from the interactions between host nuclear transcription factors and viral genes, the latter of which are believed to exist on a DNA fragment integrated into the host cells [5]. In this study, we performed whole-genome sequencing of MbBV, and based on the computational bioinformatics analysis of its genome, 116 genes were identified, 28 of which belong to the ankyrin-repeat family. Only three viral IκB-like genes among the 13 screened genes were expressed in bracovirus-induced apoptotic hemocytes. Consequently, host NF-κB/IκB, relish, dorsal, and cactus genes, and viral IκB-like vank86, vank92, and vank101 genes were each expressed in apoptosis-induced cells in a time-dependent manner. Our results suggest that viral IκB-like gene transcription did not inhibit the transcription of host NF-κB/IκB genes, suggesting that transcription of these host and viral genes have different mechanisms of regulation.
Our genomic structure analysis, which was based on Illumina Hiseq 2000 data, revealed that MbBV contains 17 potential circular dsDNA molecules of 2-20 kbp. These findings update previous expectations that MbBV possibly contains 11 circular dsDNAs with size ranges from 8-50 kbp, which were based on results obtained from gel electrophoresis [26]. In the MbBV genome, a total of 116 genes (Table 3) were identified and the existence of at least nine protein families was confirmed; furthermore, the data are mostly consistent with the data from our previously published transcriptome study, with the exception of the C-type lectin family [20]. Twenty-eight vank genes in the ORF sequence belonged to the ankyrin-repeat family, which is one of the largest gene families in MbBV. We also identified PTP in MbBV, and likewise in the congeneric MdBV, PTP and ankyrin-repeat proteins were also two of the largest gene families [40].
Table 1 shows the 13 genes screened by comparing 116 genes from the MbBV genome with the transcriptional data from hemocytes in which apoptosis had been induced by natural parasitism. The genes with mRNA expression confirmed in apoptotic hemocytes of the host on day 5 p.p. encoded six protein domains, including ankyrin-repeat, PTP, HP, BEN, Mucin-like and EGF domains, but not the RT family. One explanation for the differences observed in the transcriptional data for the genomic genes is that tissue-dependent and time-dependent transcriptional variations exist in the MbBV polydnavirus. One example of this is the eight previously identified CvBV-IκBs that have different tissue- and time-dependent transcriptional patterns [4]. Furthermore, these transcriptional patterns are consistent with our results, where virus-induced apoptotic hemocytes revealed the nature of parasitism in a short time scale of infection in the virus-induced pre-apoptosis endogenous Spli221 cell line infected by MbBV particles, as shown by the different transcription patterns for vank86, vank92, and vank101. Three vank genes were expressed in hemocytes, but not all of them were expressed in endogenous Spli221 cells; Spli221 cells are derived from the pupal ovaries of S. litura [29] and vank92 was not detected in them. At this point, a question arises about whether the PDV genome had integrated into the host cell DNA. Concerning integrated DNA fragments, Beck et al. confirmed that MdBV fragments C and J can integrate into the CiE1 cell line, which is a Pseudoplusia includens hemocyte-derived cell line [5]. As shown in Table 2, MbBV-C14, which contains vank86, mapped to MdBV fragment J, while MbBV-C16, which contains vank101, mapped to MdBV fragment I. We propose that the two contigs, NODE_25_length_3156_cov_689.371033_refined (which contains vank86) and NODE_18_length_8132_cov_284.994476_refined (which contains vank101), but not NODE_49_length_4897_cov_732.217712_refined (which contains vank92, is distributed in MbBV-C10, and maps to MdBV fragment N) can integrate into endogenous Spli221 cells over a short time scale of infection; however, the transcriptional control region still needs further exploration.
Upon parasitism, certain host genes are downregulated, while others are upregulated. In S. litura larvae infected with MbBV, 2,441 genes were downregulated, and 299 genes were upregulated; these included viral genes at 5 days p.p. [20]. These results raise a very interesting question: does viral ankyrin inhibit host NF-κB/IκB and thereby reduce host gene expression? To address this question, we identified three NF-κB/IκB factors from the host (Fig. 3A). These factors have high sequence similarities to IκB Cactus in Drosophila and were initially found to inhibit Dorsal [18]. Viral ankyrin proteins appear on the same branch of the phylogenetic tree as Cactus, which is responsible for Dorsal inhibition. Relish is regulated by an upstream molecular IκB kinase [13, 31]. Chinchore et al. reported that Relish regulates cell death in retinal degeneration in Drosophila and that this involved activation of its N-terminal domains as a toxin, suggesting that the activated form of Relish is related to the cell apoptosis pathway [9]. Additionally, in a Drosophila model of ataxia-telangiectasia, constitutively activated Relish is found to be necessary for neurodegeneration [32]. At this point, polydnaviruses may trigger the activation of Relish, but further assays may need to be performed to confirm this possibility. In Drosophila, a loss-of-function mutant of the COP9 signalosome subunit 5 causes the co-localization of Cactus and Dorsal to the nucleus and represses Dorsal-dependent transcriptional activity [18]. In alliance with Cactus, PDV might not directly affect activated Cactus, and our data support this point. There have been several reports of PDV-regulated host NF-κB/IκB at the protein level, but here, we focused on mRNA levels during transcription. In fact, the persistent expression of both proteins, viral IκB-like and host NF-κB/IκB, suggested that the viral IκB-like protein could not inhibit the transcription of NF-κB/IκB.
Normally, we would consider that the viral IκB-like protein hijacks host NF-κB to express its own genes in much the same way that other viruses do. Examples of host NF-κB participation in the viral transcription process include herpesvirus (human cytomegalovirus, HCMV) (dsDNA), papillomavirus (human papillomavirus type 16) (dsDNA), polyomavirus (simian virus 40, SV40) (dsDNA), and retrovirus (HIV type I) (ssRNA: dsDNA form integrates into the host genome). NF-κB is utilized to enhance the transcription of viral genes [15]. Unexpectedly, the same transcriptional patterns were not found, and the IκB-like protein and NF-κB/IκB both displayed time-dependent transcriptional profiles; thus, the transcription of the viral IκB-like protein may involve other transcriptional factors, which still await identification. Given that viral IκB-like genes are potentially integrated into DNA fragments in the host cell, it will be interesting to investigate the interaction between the viral promoter of the integrated DNA fragments and nuclear transcriptional factors in the host. Naturally, we believe that polydnaviruses as gene delivery systems will be useful genome editing tools in the future.
Abbreviations
- MbBV:
-
Microplitis bicoloratus bracovirus
- MdBV:
-
Microplitis demolitor bracovirus
- CcBV:
-
Cotesia congregata bracovirus
- CsBV:
-
Cotesia sesamiae bracovirus
- CvBV:
-
Cotesia vestalis bracovirus
- CiBV:
-
Chelonus inanitus bracovirus
- PDV:
-
polydnavirus
- Vank:
-
viral ankyrin
References
Albrecht U, Wyler T, Pfister-Wilhelm R, Gruber A, Stettler P, Heiniger P, Kurt E, Schiimperli D, Lanzrein B (1994) Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J Gen Virol 75:3353–3363
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome biology 11:R106
Annaheim M, Lanzrein B (2007) Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J Gen Virol 88:450–457
Bae S, Kim Y (2009) IkB genes encoded in Cotesia plutellae bracovirus suppress an antiviral response and enhance baculovirus pathogenicity against the diamondback moth, Plutella xylostella. J Invertebr Pathol 102:79–87
Beck MH, Zhang S, Bitra K, Burke GR, Strand MR (2011) The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens. J Virol 85:11685–11696
Beckage NE (2008) Parasitoid Polydnaviruses and Insect Immunity. In: Beckage NE (ed) Insect Immunology. Academic Press, San Diego, p 243
Bitra K, Suderman RJ, Strand MR (2012) Polydnavirus Ank proteins bind NF-kappaB homodimers and inhibit processing of Relish. PLoS Pathog 8:e1002722
Chen YF, Gao F, Ye XQ, Wei SJ, Shi M, Zheng HJ, Chen XX (2011) Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology 414:42–50
Chinchore Y, Gerber GF, Dolph PJ (2012) Alternative pathway of cell death in Drosophila mediated by NF-kappaB transcription factor Relish. Proc Natl Acad Sci U S A 109:E605–E612
Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fuester RW, Schatz MC, Pedroni MJ, Fadrosh DW, Haas BJ, Toms BS, Chen D, Nene V (2007) Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus. BMC Microbiol 7:61
Desjardins CA, Gundersen-Rindal DE, Hostetler JB, Tallon LJ, Fadrosh DW, Fuester RW, Pedroni MJ, Haas BJ, Schatz MC, Jones KM, Crabtree J, Forberger H, Nene V (2008) Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome biology 9:R183
Dupuy C, Gundersen-Rindal D, Cusson M (2012) Genomics and Replication of Polydnaviruses. In: NEB-M D (ed) Parasitoid viruses. Academic Press, San Diego, pp 47–61
Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, Silverman N (2009) Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA 106:9779–9784
Espagne E, Dupuy C, Huguet E, Cattolico L, Provost B, Martins N, Poirie M, Periquet G, Drezen JM (2004) Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306:286–289
Flint SJ, Racaniello VR, Enquist LW, Skalka AM (2009) Principles of Virology. Molecular Biology. American Society of Microbiology, Washington, DC
Flores-Saaib RD, Jia S, Courey AJ (2001) Activation and repression by the C-terminal domain of dorsal. Development, pp 1869–1879
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235
Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, Segal D, Chamovitz DA (2007) COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: effects on Cactus, Dorsal and hematopoiesis. Genes Cells 12:183–195
Li G, Chen Q, Pang Y (1998) Studies of artificial diets for the beet armyworm, Spodoptera exigua. Acta Sci Circumstant/Huanjing Kexue Xuebao 4:1–5
Li M, Pang Z, Xiao W, Liu X, Zhang Y, Yu D, Yang M, Yang Y, Hu JS, Luo KJ (2014) A transcriptome analysis suggests apoptosis-related signaling pathways in hemocytes of Spodoptera litura after parasitization by Microplitis bicoloratus. PLoS One 9:e110967
Liu T, Li M, Zhang Y, Pang Z, Xiao W, Yang Y, Luo KJ (2013) A role for innexin2 and innexin3 proteins from Spodoptera litura in apoptosis. PLoS One 8:e70456
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Loytynoja A, Goldman N (2010) webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinform 11
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Luo KJ, Pang Y (2006) Spodoptera litura multicapsid nucleopolyhedrovirus inhibits Microplitis bicoloratus polydnavirus-induced host granulocytes apoptosis. J Insect Physiol 52:795–806
Luo KJ, Pang Y (2006) Disruption effect of Microplitis bicoloratus polydnavirus EGF-like protein, MbCRP, on actin cytoskeleton in lepidopteran insect hemocytes. Acta Biochim Biophys Sin (Shanghai) 38:577–585
Luo KJ, Trumble JT, Pang Y (2007) Development of Microplitis bicoloratus on Spodoptera litura and implications for biological control. BioControl 52:309–321
Michaely P, Tomchick DR, Machius M, Anderson RG (2002) Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J 21:6387–6396
Mitsuhashi J (1995) A continuous cell line from pupal ovaries of thecommon cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Appl Entomol Zool 30:75–82
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Park JM, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, Kato T Jr, Richards N, Chan K, Mercurio F, Karin M, Wasserman SA (2004) Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev 18:584–594
Petersen AJ, Katzenberger RJ, Wassarman DA (2013) The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics 194:133–142
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374–378
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123
Stoltz D, Lapointe R, Makkay A, Cusson M (2007) Exposure of ichnovirus particles to digitonin leads to enhanced infectivity and induces fusion from without in an in vitro model system. J Gen Virol 88:2977–2984
Strand MR (1994) Microplitis Demolitor polydnavirus infects and expresses in specific morphotypes of Pseudoplusia Includens haemocytes. J Gen Virol 75:3007–3020
Strand MR, Burke GR (2012) Polydnaviruses as symbionts and gene delivery systems. PLoS Path 8. doi:10.1372/journal.ppat.1002757
Thoetkiattikul H, Beck MH, Strand MR (2005) Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc Natl Acad Sci USA 102:11426–11431
Tian SP, Zhang JH, Wang CZ (2007) Cloning and characterization of two Campoletis chlorideae ichnovirus vankyrin genes expressed in parasitized host Helicoverpa armigera. J Insect Physiol 53:699–707
Webb BA, Strand MR, Dickey SE, Beck MH, Hilgarth RS, Barney WE, Kadash K, Kroemer JA, Lindstrom KG, Rattanadechakul W, Shelby KS, Thoetkiattikul H, Turnbull MW, Witherell RA (2006) Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347:160–174
Yanase T, Yasunaga C, Kawarabata T (1998) Replication of Spodoptera exigua nucleopolyhedrovirus in permissive and non-permissive lepidopteran cell lines. Acta Virol 42:293–298
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Acknowledgments
The authors are grateful to Dr. Yonggyun Kim (Andong University, Korea) and Dr. Sandra Cheesman for proofreading this manuscript. The authors are grateful to Dr. Kai Zhou (University Medical Center Groningen, The Netherlands) for submitting the DNA sequences to GenBank. This work was supported in part by a grant (2013CB127600) from National Basic Research Program of China, grants from National Natural Science Foundation of China (31260448; 31060251) and a grant from Yunnan Department of Science and Technology (2013FA003) to K.L.; M.L. was supported by a grant from National Natural Science Foundation of China (31360454) and Yunnan Department of Science and Technology (2012FB120).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
D.-S. Yu, Y.-B. Chen and M. Li are equal contributors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, DS., Chen, YB., Li, M. et al. A polydnaviral genome of Microplitis bicoloratus bracovirus and molecular interactions between the host and virus involved in NF-κB signaling. Arch Virol 161, 3095–3124 (2016). https://doi.org/10.1007/s00705-016-2988-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2988-3