Abstract
We report the first complete genome sequence of tropical soda apple mosaic virus (TSAMV), a tobamovirus originally isolated from tropical soda apple (Solanum viarum) collected in Okeechobee, Florida. The complete genome of TSAMV is 6,350 nucleotides long and contains four open reading frames encoding the following proteins: i) 126-kDa methyltransferase/helicase (3354 nt), ii) 183-kDa polymerase (4839 nt), iii) movement protein (771 nt) and iv) coat protein (483 nt). The complete genome sequence of TSAMV shares 80.4 % nucleotide sequence identity with pepper mild mottle virus (PMMoV) and 71.2-74.2 % identity with other tobamoviruses naturally infecting members of the Solanaceae plant family. Phylogenetic analysis of the deduced amino acid sequences of the 126-kDa and 183-kDa proteins and the complete genome sequence place TSAMV in a subcluster with PMMoV within the Solanaceae-infecting subgroup of tobamoviruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus Tobamovirus of the family Virgaviridae is composed of 35 species [1]. Tobamoviruses are characterized by having rigid rod-shaped virions and single-stranded positive-sense RNA genomes. These viruses are easily transmitted by mechanical inoculation and plant-to-plant contact and are distributed worldwide [2]. Genomes of all characterized tobamoviruses contain four open reading frames encoding the following proteins: a 124- to 132-kDa replicase protein with methyltransferase and RNA helicase domains, a 181- to 189-kDa replicase protein with a polymerase domain, a movement protein (MP), and a coat protein (CP) [2, 7, 8].
Tropical soda apple (Solanum viarum Dunal) is a noxious solanaceous weed native to South America [9] that has spread throughout Florida and the southern United States since being introduced in the 1980s [4, 5]. Tropical soda apple mosaic virus (TSAMV) was originally isolated from tropical soda apple plants displaying foliar mosaic symptoms that were collected in Okeechobee, FL [3]. Host range analysis of TSAMV demonstrated that it infected 16 species of plants in the family Solanaceae. Molecular and phylogenetic analysis of TSAMV MP and CP indicated high sequence similarity to pepper mild mottle virus (PMMoV) and other Solanaceae-infecting tobamoviruses [3]. However, the complete genome sequence of TSAMV has not been reported.
The original Nicotiana tabacum cv Xanthi nc local-lesion-passaged TSAMV isolate [3] was used to inoculate Nicotiana benthamiana, from which virions were partially purified from systemically infected tissue using a typical tobamovirus protocol [13]. TSAMV genomic RNA extraction, RNA-seq library preparation, and sequencing on an Ion Torrent Personal Genome Machine (Life Technologies) were as described previously [6]. The complete genome consensus sequence was generated using the Torrent Mapping Alignment Program (TMAP version 4.21), Geneious R8 version 8.1.4 (Biomatter, Auckland, New Zealand) and Integrative Genomic Viewer (IGV) version 2.3 [10, 12]. MEGA6 was used for multiple sequence alignments of the nucleotide and deduced amino acid sequences, and for phylogenetic analysis [11]. Pairwise comparisons of TSAMV and other tobamoviruses were made with DNA Master (available from http://cobamide2.bio.pitt.edu/).
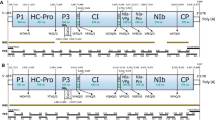
The complete genome of TSAMV (KU659022) is 6,350 nucleotides (nt) long and includes a 5′ untranslated region (UTR, nt 1-69), genes encoding a 183-kDa polymerase (nt 70-4908), 126-kDa methyltransferase/helicase (nt 70-3423), MP (nt 4910-5680), and CP (nt 5683-6165), and a 3′ UTR (nt 6166-6350) (Fig. 1A). Comparison of the TSAMV 126-kDa, 183-kDa, MP and CP amino acid sequences with those of other tobamoviruses showed sequence identities ranging from 40.2 to 90.2 %, 45.0 to 90.1 %, 32.2 to 80.9 % and 37.4 to 84.8 %, respectively (Table 1). Both the 126-kDa and 183-kDa proteins showed highest amino acid identity (90.2 % and 90.1 %, respectively) to the corresponding proteins of PMMoV, consistent with previous sequence analysis of the TSAMV MP and CP [3]. At the complete-genome level, TSAMV shared 80.4 % sequence identity with PMMoV. When comparing the TSAMV genome sequence with available complete genome sequences of 28 other tobamoviruses, the closest nucleotide sequence identities (71.2-80.4 %) were with other Solanaceae-infecting tobamoviruses.
Genome organization (A) and phylogenetic analysis of TSAMV and recognized members of the genus Tobamovirus based on the deduced amino acid sequences of the 124- to 132-kDa protein (B), 181- to 189-kDa protein (C), and complete genome sequence (D). Genomic RNA is shown as a solid line, with nucleotides indicated at 5′ and 3′ termini. Predicted open reading frames are shown as boxes containing protein names and nucleotides (shown above or below) indicating start and stop codons. Unrooted, neighbor-joining trees were inferred using MEGA6 [11], with tobacco rattle virus (TRV; NP_620669.1 and NP_620669.2) as an outgroup in B and C. Branch lengths indicate the number of amino acid or nucleotide differences per site, and the numbers at the nodes indicate bootstrap values greater than 70 based on bootstrap analysis of 1000 replicates. The following viruses were included (listed with abbreviation and GenBank accession no. for nucleotide sequence): bell pepper mottle virus (BpeMV, NC_009642), brugmansia mild mottle virus (BrMMV, NC_010944), cactus mild mosaic virus (CMMoV, NC_011803), clitoria yellow mottle virus (CIYMV, NC_016519), cucumber fruit mottle mosaic virus (CFMMV, NC_002633), cucumber green mottle mosaic virus (CGMMV, NC_001801), cucumber mottle virus (CMoV, NC_008614), frangipani mosaic virus (FrMV, NC_014546), hibiscus latent Fort Pierce virus (HLFPV, NC_025381.1), hibiscus latent Singapore virus (HLSV, NC_008310.2), kyuri green mottle mosaic virus (KGMMV, NC_003610.1), maracuja mosaic virus (MarMV, NC_008716), Obuda pepper virus (ObPV, NC_003852), odontoglossum ringspot virus (ORSV, NC_001728), paprika mild mottle virus (PaMMV, NC_004106), passion fruit mosaic virus (PafMV, NC_015552) pepper mild mottle virus (PMMoV, NC_003630), rehmannia mosaic virus (RheMV, AB628188), ribgrass mosaic virus (RMV, NC_002792.2), streptocarpus flower break virus (SFBV, NC_008365), tobacco mild green mosaic virus (TMGMV, NC_001556), tobacco mosaic virus (TMV, NC_001367), tomato mosaic virus (ToMV, KR537870.1), tropical soda apple mosaic virus (TSAMV, KU659022), turnip vein clearing virus (TVCV, NC_001873), wasabi mottle virus (WMoV, NC_003355), yellow tailflower mild mottle virus (YTMMV, KF495564), youcai mosaic virus (YoMV, NC_004422), and zucchini green mottle mosaic virus (ZGMMV, NC_003878). Solanaceae-, Brassicaceae- and Cucurbitaceae-infecting subgroups are indicated
Phylogenetic analysis of the deduced amino acid sequences of the 126-kDa and 183-kDa proteins (Fig. 1B and C), and the complete genome sequence (Fig. 1D), revealed the previously demonstrated clusters of the Solanaceae-, Brassicaceae- and Cucurbitaceae-infecting tobamovirus subgroups [2, 3, 7]. In all cases, TSAMV was placed in a subcluster with PMMoV within the Solanaceae-infecting subgroup of tobamoviruses, confirming that TSAMV is most closely related to, but distinct from, PMMoV.
References
Adams MJ, Antoniw JF, Kreuze J (2009) Virgaviridae: a new family of rod-shaped plant viruses. Arch Virol 154:1967–1972
Adams MJ, Heinze C, Jackson AO, Kreuze JF, Macfarlane SA, Torrance L (2012) Tobamovirus. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier/Academic Press, London, pp 1153–1156
Adkins S, Kamenova I, Rosskopf EN, Lewandowski DJ (2007) Identification and characterization of a novel tobamovirus from tropical soda apple in Florida. Plant Dis 91:287–293
Coile NC (1993) Tropical soda apple, Solanum viarum Dunal: the plant from hell. Florida Department of Agriculture and Consumer Services, Division of Plant Industry. Bot Circ 27
Ferrell JA, Mullahey JJ (2005) Tropical soda apple (Solanum viarum Dunal) in Florida. Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida, Gainesville. Publ. #SS-AGR-50
Fillmer K, Adkins S, Pongam P, D’Elia T (2015) Complete genome sequence of a tomato mottle mosaic virus isolate from the United States. Genome Announc 3:e00167-15
Gibbs A (1999) Evolution and origin of tobamoviruses. Philos Trans R Soc Lond B 354:593–602
Lewandowski DJ, Dawson WO (2000) Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90–98
Nee M (1991) Synopsis of Solanum section Acanthophora: a group of interest for glycol-alkaloids. In: Hawkes JW, Lester RN, Nee M, Estrada N (eds) Solanaceae III: taxonomy, chemistry, evolution. Royal Botanic Gardens, Surrey, pp 258–266
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192
Wetter C, Conti M (1988) Pepper mild mottle virus. CMI/AAB descriptions of plant viruses no. 330
Acknowledgments
We thank Carrie Vanderspool, Megan Carroll and Yaowapa Tantiwanich for their excellent technical assistance. Financial support was provided in part by The Banack Family Partnership Endowed Teaching Chairs in Agriculture to P.P. and T.D.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Fillmer, K., Adkins, S., Pongam, P. et al. The complete nucleotide sequence and genomic characterization of tropical soda apple mosaic virus. Arch Virol 161, 2317–2320 (2016). https://doi.org/10.1007/s00705-016-2888-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2888-6