Abstract
Crimean-Congo haemorrhagic fever (CCHF) is a potentially fatal systemic viral disease in many parts of the world, including Iran. The nationwide incidence of human CCHF in endemic areas was 870 confirmed cases with 126 deaths (case fatality rate, CFR = 17.6 %) in the decade leading to 2012. The detection of the CCHF virus (CCHFV) genome in tick vectors is of fundamental importance for identifying these ticks as potential reservoirs of CCHFV infection. From May to October 2013, following detection of four new clinical cases resulting in two deaths in the city of Mashhad (northeast Iran), hard ticks were recovered from infested livestock in 40 villages in Khorasan-Razavi province and examined by the microscopic method for species identification. About a quarter of the ticks were then subjected to reverse-transcription polymerase chain reaction (RT-PCR) to detect the CCHFV genome. The PCR products were then sequenced, and their phylogenetic lineages were determined. A total of 407 hard ticks were captured, representing seven different species in two distinct genera. Members of the genus Hyalomma were widely distributed in all but two of the villages studied, and this was also the most frequent (83.3 %) tick genus. Of 105 adult ticks subjected to RT-PCR, four (3.8 %) ticks were found positive for the CCHFV genome. One brown ear tick, Rhipicephalus appendiculatus, was found to be naturally infected for the first time anywhere in the world. Ticks of Hyalomma asiaticum, Hyalomma marginatum, and Rhipicephalus turanicus were also found to be naturally infected with CCHFV. CCHFV found in these four different tick species were clustered in the same lineage with the Matin and SR3 strains from Pakistan and some other strains from Iran, indicating that these tick species were naturally infected with genetically closely related CCHFV in the region. The presence of CCHFV infection in four different hard tick species was confirmed using RT-PCR in northeast Iran. Part of this infection was attributed to Rh. appendiculatus, which is thus a potential new natural vector of CCHFV in Iran. It is also confirmed by phylogenetic analysis that CCHFV in this region is genetically closely related, even in the different tick species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crimean-Congo hemorrhagic fever (CCHF) is caused by members of the genus Nairovirus in the family Bunyaviridae [1]. Despite being an enzootic infection, outbreaks and sporadic small clusters of disease cases occur focally in humans after direct bites of infected ticks, which are competent natural reservoirs, or via skin contact with viremic tissues, blood, or other contaminated body fluids of patients or infectious asymptomatic domestic and wild livestock such as cattle, camels, sheep and goats [2, 3]. Although CCHF virus (CCHFV) is transmitted to a wide variety of animals, it is severely pathogenic only for humans [4]. The large burden of human CCHF cases in Iran and neighbouring countries (≈5000 patients in the first decade of this century) points to the serious importance of this challenge [5].
Specific antibodies against CCHFV antigens in the sera of humans, cattle and sheep were first demonstrated back in the early 1970s [6]. Current molecular epidemiology and phylogenetic data indicate that the widest genetic diversity of CCHFV in any country occurs in Iran, where there are at least four (genotype 1 [Africa 1]; genotype 2 [Asia 2]; genotype 4 [Europe 1]; genotype 7 [Africa 1]) or five different circulating genomic variants out of a total of seven distinct genotypes [7–9].
Viruses were isolated from hard (ixodid) ticks [10]. The first human cases were seen in 1999 and have since been recorded in all but four (87 %) Iranian provinces [8, 11]. Nosocomial infection with CCHFV is also being reported, with immense and alarming implications [11]. As a result of a recent hospital outbreak of CCHF in the city of Mashhad, Khorasan-Razavi province in northeast Iran, four patients were hospitalized, two of whom (one of them a nurse) died later due to clinical complications [11].
Ticks are one of the most medically important groups of bloodsucking arthropods, which may carry a wide variety of pathogens responsible for many tropical zoonotic diseases, including those caused by the arboviruses in the genus Nairovirus. The genus of hard tick, Rhipicephalus, includes numerous species that are important vectors of infectious agents. Rhipicephalus appendiculatus (known as the “brown ear tick”) is a three-host bloodsucking inornate species of hard tick.
This study describes the first identification of the CCHFV genome in the Rh. appendiculatus species of hard tick in northeastern Iran. While CCHFV has been experimentally shown to infect members of this species, [12] and other species of Rhipicephalus have been shown to be infected in nature, this is the first demonstration of naturally occurring infection in this particular species of Rhipicephalus tick. While this may then appear to be an incremental increase in understanding, the identification of additional tick species that are able to serve as vectors for CCHFV in the endemic areas is an important and neglected area of research with significant public health implications for the transmission dynamics of this disease.
This outbreak of CCHF prompted us to conduct research on faunal identification of hard ticks feeding on livestock and their likely infection with CCHFV using a reverse transcription polymerase chain reaction (RT-PCR) method. To the best of our knowledge, this is the first global report on natural detection of CCHFV in a brown ear hard tick, Rh. appendiculatus, in northeastern Iran. The natural presence of these viruses in wild-caught Rh. appendiculatus has never been reported before.
Materials and methods
Study area
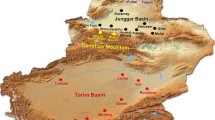
Khorasan-Razavi province is located on the northeastern frontier of Iran (Fig. 1). This study focused on the three counties of Torbat-Heydariyeh (35°15′N, 59°132′E at 1333 m above sea level), Roshtkhar (34°58′N, 59°38′E at 1215 m a.s.l.), and Khaf (34°38′N, 60°10′E at 970 m a.s.l.). For the present study, 15 villages each from Torbat-Heydariyeh and Roshtkhar, and 10 villages from Khaf that each had at least 10 households, were randomly selected.
Collection of ticks
Hard ticks were collected from infested livestock for six months from May to October 2013, when peak densities of ticks were expected [13]. Adult ticks were collected from different parts of the bodies of sheep, goats and cows by clustered random sampling in the 40 sampled villages. The exclusion criteria were recently insecticide-sprayed animal husbandry units and those that did not consent to sampling. The whole body of each ruminant host was examined for the presence of tick infestation by palpation, principally on the ears and along the nape of the neck, perineum, scrotum or udder, and tail base. With the aid of experienced animal breeders, 3 hours were spent searching for ticks on infested animals at each station. All ticks were handled with sterile gloves and fine forceps. They were identified at the species level by trained acarologists. Each tick was cautiously dislodged and transferred alive into a separate screw-capped sterile vial with a small piece of cotton soaked in 1 % mycostatin solution to inhibit desiccation and mold growth. Each vial was labeled accordingly, frozen at -40 °C, and subsequently, only depleted (non-engorged) adult ticks were checked for the CCHF virus genome at Pasteur Institute of Tehran, Iran.
RNA extraction from ticks
Each of 105 adult ticks, chosen as a random sample of the collected ticks, was separately washed twice with phosphate-buffered saline (PBS, pH = 7.4) and crushed with a pestle and mortar in 300 µL of PBS. Viral RNA was extracted from 140 µL of phenol-extracted tick suspensions using a QIAamp RNA Mini kit (QIAGEN GmbH, Hilden, Germany) according to the instructions of the manufacturer.
RT-PCR
Reverse transcription of RNA was conducted using previously described primers [14] that amplify a 536-bp fragment of the S segment of the CCHF viral genome. Briefly, the extracted viral RNA was analyzed by gel-based and real-time RT-PCR using a one-step RT-PCR kit (QIAGEN) with specific primers: F2 primer (5′-TGGACACCTTCACAAACTC-3′) and R3 primer (5′-GACAAATTCCCTGCACCA-3′) [15]. The PCR reaction was done in a total volume of 50 µL. The reaction conditions were 30 min at 50 °C, 15 min at 95 °C, 40 cycles of 30 s at 95 °C, 30 s at 50 °C, and 45 s at 72 °C, and finally, 10 min at 72 °C as a final extension step. For qualitative analysis, 1 µL of SYBR green dye at 1:10,000 dilution was also used in the master mix. For gel-based analysis, 5 µL of the PCR products were mixed with 1 µL of loading buffer, and electrophoresis was carried out in a 2 % agarose gel in Tris-borate EDTA buffer (TBE). DNA bands were stained with ethidium bromide and were visualized on a UV transilluminator.
Sequencing and gene sequence submission
Amplified PCR products were sequenced directly by a modified Sanger sequencing method using an ABI 3130 Genetic Analyzer and a BigDye terminator cycle sequencing reaction kit V3.1. Four new sequences (Nad-8-494c, Hy. asiaticum; Nad-20-510c, Hy. marginatum; Nad-71-496c, Rh. appendiculatus; and Nad-83-519c, Rh. turanicus) were submitted to GenBank and assigned the accession numbers KP223318, KP223319, KP223320 and KP223321, respectively.
Phylogenetic and sequence analysis
The sequences of the CCHFV amplicons from this study and 44 sequences retrieved from GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) (Table 3) were aligned using the Clustal W Multiple Sequence Alignment (MSA) tool with the following parameters: gap opening penalty, 15; gap extension penalty, 6.66; DNA weight matrix IUB and transition weight, 0.5.
A phylogenetic tree was constructed by maximum-likelihood statistical method using the Kimura 2-parameter model. The reliability of the phylogenetic tree was evaluated using the bootstrap test based on 1000 replicates at a cutoff value of 70 %. Pairwise distances were computed using MEGA 5 software. Phylogenetic and sequence analyses were conducted using MEGA version 5 [16].
Results
A total of 407 adult ticks were collected from the three counties of Torbat-Heydaryieh (160 ticks, 39.3 %), Roshtkhar (144 ticks, 35.4 %) and Khaf (103 ticks, 25.3 %) during the study period. All the villages in this study were infested with no fewer than three and no more than 17 different tick specimens (Table 1). Only six villages among the 40 study stations, including the village of Safiabad in the county of Torbat-Heydaryieh, had no more than six ticks in their sampling collections.
At Torbat-Heydaryieh, different numbers of tick species were removed from cattle (71 ticks, 44.3 %), sheep (57 ticks, 35.6 %), and goats (32 ticks, 20 %). At Roshtkhar, multiple tick species were isolated from cattle (18 ticks, 12.5 %), sheep (99 ticks, 68.7 %), and goats (27 ticks, 18.8 %). At Khaf, these values were 43 (41.8 %), 44 (42.7 %) and 16 ticks (15.5 %), respectively.
Two distinct tick genera were represented, from which seven different tick species (three Hyalomma and four Rhipicephalus) were identified on infested livestock in the three counties of Khorasan-Razavi province that were studied (Table 1). Ticks of the genus Hyalomma was found to be the most widely distributed in all but two of the villages. It was also the most frequent (83.3 %) tick genus. The least abundant tick species (1 tick, 0.24 %) was Hy. anatolicum, which was found in only one village, while the least common (3 ticks, 0.74 %) tick species in the genus Rhipicephalus was the brown dog tick Rh. sanguineus. The most abundant species were Hy. asiaticum and Hy. marginatum, with 66.6 % and 18.5 %, respectively. The latter species was present in only half of the villages surveyed, whereas the largest number of Hy. marginatum was recorded in a village known as Ciuki in Torbat-Heydaryieh county. Although both tick species (asiaticum and marginatum) were found to be widely distributed in the surveyed region, neither of them was present in all sampled locations in the study area.
The other tick genus was Rhipicephalus, which was represented by four different species comprising only 16.7 % of the total sample size. The first and second most abundant species in the genus of Rhipicephalus were Rh. bursa (34 ticks, 8.3 %) and Rh. turanicus (26 ticks, 6.4 %), respectively. The other two rarely found species were the brown dog tick, Rh. sanguineus, referred to above, and the brown ear tick, Rh. appendiculatus. Only five (1.2 %) specimens of the latter species were found in the villages of Safiabad and Janatabad.
Using reverse transcription PCR on a selection of collected ticks (~26 %), it was revealed that a small number (4, 3.8 %) of ticks from both genera, Hyalomma and Rhipicephalus, removed from infested hosts were naturally infected with CCHFV (Table 2). A total of 105 depleted adult male and female ticks from two distinct genera were included in the molecular assay in this study. Most of the selected ticks were of the species Hy. asiaticum (58 ticks, 55.2 %) and Hy. marginatum (29 ticks, 27.6 %). The number of Hy. anatolicum, Rh. bursa, Rh. turanicus, Rh. sanguineus, and Rh. appendiculatus ticks identified was 1 (0.95 %), 7 (6.7 %), 5 (4.8 %), 1 (0.95 %), and 4 (3.8 %), respectively.
All of the selected ticks were subjected to nested RT-PCR. The natural presence of the CCHFV genome was only detected in one sample each of Hy. asiaticum (1.7 %), Hy. marginatum (3.4 %), Rh. turanicus (20 %), and Rh. appendiculatus (25 %) (Fig. 2). The positivity rate in Rhipicephalus ticks was 12 %, and that of Hyalomma ticks was about 2 %. No Hy. anatolicum, Rh. bursa or Rh. sanguineus ticks gave positive nested RT-PCR results for CCHF virus.
Current molecular epidemiology and phylogenetic data have indicated the existence of seven distinct clusters of CCHFV (Tables 3 and 4). All four of the newly found strains of CCHFV identified in Hyalomma asiaticum, Hy. marginatum, Rhipicephalus turanicus, and Rh. appendiculatus in Khorasan-Razavi province of Iran were clustered in the same lineage of Clade IV (Asia 1) with Matin; and SR3 strains in Pakistan; Afg09-2990 in Afghanistan; and Iran-52, Iran-53, and 786/02 strains in Iran (Fig. 3). These four strains were genetically distinguishable from the Baghdad strain in Iraq, the Oman strain in Oman, and SCT ex Afghanistan in Afghanistan (Fig. 3). These results indicate that CCHFV in Khorasan-Razavi province of Iran is genetically closely related, even in the different tick species.
Unrooted phylogenetic tree based on a portion of the S segment (503 nucleotides) of CCHF virus, constructed by the maximum-likelihood using Mega5 software. Dugbe virus (strain KT281/75AF434165) and Hazara virus (strain JC280M86624) were used as outgroups. The new sequences from this study are shown in a box. The numbers at nodes indicate the bootstrap values for 1000 replicates. Clade I, W. Africa 1; Clade II, C. Africa; Clade III, S. Africa and/or W. Africa 2; Clade IV, Asia 1; Clade IV, Asia 2; and Clade VI, Greece AP92
Discussion
Tick-borne fevers, which are caused by a wide range of pathogens, are of increasing public-health significance in Iran [17–19]. CCHFV is a polythetic and zoonotic microparasite of engorging hard and soft ticks with a wide spectrum host specificity. This virus can be transmitted by the venereal, trans-stadial and trans-ovarial routes in ticks [20]. Infections due to viruses being transmitted via haematophagous arthropods (arthropod-borne viruses or arboviruses such as CCHFV) impose a particularly persistent and substantial burden of disease, in addition to those from non-infectious [21] or other infectious diseases [22–24] on the lives of healthy Iranians. Co-infections with these infectious agents could simultaneously be found in endemic regions [25].
The present study was conducted in a northeast Iranian Khorasan-Razavi province where a recent sero-epidemiological survey of CCHFV in slaughterhouse workers indicated an infection rate of 17.5 % [26]. This province was also shown to be one of the hot spots of CCHFV infection in livestock [27]. The national incidence of this clinical disease in endemic areas was 870 confirmed cases with 126 deaths (case fatality rate [CFR], 17.6 %) in the decade leading to the year of 2012 [9]. On the other hand, a CFR of only 5 % was reported in the northwestern neighbouring country of Turkey. The gap in CFR values of Iran and Turkey was suggested to be due to different surveillance (sporadic case reports in Iran vs. a large case series including mild disease cases in Turkey) or better laboratory diagnosis of the patients in Turkey, which also included patients with mild symptoms [1, 28]. It is also possible that the gap is partly due to the difference in supportive care operating in those adjacent countries.
The detection of CCHFV in tick vectors and confirmation of their vector competence through transovarial and venereal transmission of CCHFV is of paramount importance for determining whether they are potential reservoirs of infection [1]. However, few attempts have been made to identify tick vectors of CCHFV, despite their clinical indispensability. In a study in the west of Iran, Hamadan province, researchers found three of four, two of three, and one of three different species in the genera of Hyalomma, Rhipicephalus, and Haemaphysalis, respectively, to be positive for the CCHFV genome by RT-PCR, [29]. In a later report from the same province, both hard and soft ticks were positive for the CCHFV genome [30]. These viruses were all genetically clustered in the main strain group circulating in Iran and closely related to the Pakistani Matin strain, except one isolate from Rh. sanguineus, which showed more divergence from the other isolates, possibly correlated to its geographical range [30]. The current study also found members of two species each in the genera Hyalomma and Rhipicephalus to be positive for the CCHFV genome. Most (75 %) of the CCHFV-positive samples originated from the northernmost study area, Torbat-Heydariyeh county. In an earlier report from Kurdistan province, northwest Iran, only Hyalomma ticks were found to be infected with CCHFV. Three remaining genera of ticks were all negative in the RT-PCR assay [18].
The eco-climatic conditions in Iran and neighbouring countries are favorable for many ticks to transmit CCHFV in nature [4, 5]. Similarly, disease outbreaks have recently progressed against a background of optimal climatic parameters and ambient changes suitable for the presence of ticks and their infested hosts [31, 32]. In addition, trans-border transmission has also been suggested to be involved in CCHFV circulation [7, 33]. In these endemic regions, most tick-borne multi-host zoonotic infections of domesticated animals are treated with the use of acaricides. These infested ungulates are mostly in contact with wildlife, so transmission of the infection could hardly be prevented [34]. It has been proposed that more spatio-temporal studies are needed to elucidate the natural history of the CCHFV-tick-host system, and transmission modeling is required to better understand the maintenance of CCHFV foci in all Iranian provinces [35].
Four tick species (Hy. asiaticum, Hy. marginatum, Rh. turanicus,, Rh. appendiculatus) that were positive for the CCHFV genome were naturally caught from infested ungulates in the study region. The brown ear tick, Rh. appendiculatus, was positive for the CCHFV genome by RT-PCR. No evidence of its natural infection with the arbovirus causing CCHF has so far been provided. This is the first report of CCHFV infection of brown ear ticks anywhere in the world. Ticks of the genus Hyalomma are still considered to be the main vectors of CCHFV, but increasing reports of infection of Rhipicephalus ticks suggest a potentially supportive role in viral transmission of CCHF. In accordance with the results of previous studies conducted elsewhere in different parts of the region, [15, 36] it is speculated that Rhipicephalus spp. could play an auxiliary role in the epidemiology of CCHFV in Iran.
It is well known that Hyalomma ticks are major vectors of CCHFV, and multiple hard tick species are presumed to be involved in transmission of CCHF viruses. Brown dog ticks (Rh. sanguineus) were found to be infected with CCHFV in Bulgaria, Iran, and Turkey [30, 37, 38]. Rh. turanicus was also reported to be positive for the CCHFV genome in Turkey [36, 38, 39]. Rh. bursa was positive in Turkey and Greece [38–40]. Rh. guilhoni and Rh. evertsi evertsi in Senegal and Rh. evertsi mimeticus in Africa were also reported to have the CCHFV genome [41, 42]. The new evidence of a CCHFV genome in Rh. appendiculatus is thus an important observation that needs to be considered in any novel reappraisal of the molecular epidemiology and transmission dynamics of CCHFV in southwest Asia.
All four strains of CCHFV identified in Hyalomma asiaticum, Hy. marginatum, Rh. turanicus, and Rh. appendiculatus in Khorasan-Razavi province were clustered in the same lineage with the Matin and SR3 strains from Pakistan; Afg09-2990 from Afghanistan; and the Iran-52, Iran-53, and 786/02 strains from Iran (Fig. 3). These four strains were genetically distinguishable from the Baghdad strain from Iraq, the Oman strain from Oman, and the SCT ex Afghanistan strain from Afghanistan (Fig. 3). These results showed that CCHFV in this province of Iran is genetically closely related, even in the different tick species. This finding is in line with other reports [8, 30, 43]. However, the Oman and Iraq lineages showed higher divergence from the virus sequences in the present study.
There is no doubt that a single RT-PCR-positive finding in a tick species does not necessarily equate to its status as a competent vector. Since the tick samples selected for RT-PCR were depleted, this rules out the possibility of ingestion of a CCHFV-positive blood meal from an infected ungulate host without actual infection of the tick itself. Furthermore, even if a tick species is legitimately CCHFV positive, this does not mean that it is competent as a vector. It should be noted that several other criteria have to be met in order for a given arthropod species to have status of vector. Some of these criteria are (1) natural bloodsucking of the suspected tick on humans or other hosts, (2) sustained survival of the virus at different life cycle stages (eggs, larvae, nymphs) of the vector, (3) exact identity of the virus in the vector and vertebrate host, (4) the ability of the vector to transmit the virus by bite, and (5) vector abundance.
In conclusion, the presence of CCHFV in four different species of hard ticks in northeast Iran was confirmed using RT-PCR. For the first time, the viral genome was found in the brown ear tick Rh. appendiculatus, suggesting that it is a potential new natural vector of CCHFV in Iran. It was also shown by phylogenetic analysis that CCHFV isolates in this region are genetically closely related, even in ticks of different species. They were clustered in the same lineage with some strains from Afghanistan, Iran and Pakistan.
References
Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M (2013) Crimean-Congo hemorrahgic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res 100:159–189
Papa A, Velo E, Papadimitriou E, Cahani G, Kota M, Bino S (2009) Ecology of the Crimean-Congo hemorrhagic fever endemic area in Albania. Vect Zoonotic Dis 9:713–716
Champour M, Mohammadi G, Chinikar S, Razmi G, Shah-Hosseini N, Khakifirouz S, Mostafavi E, Jalali T (2014) Seroepidemiology of Crimean-Congo hemorrhagic fever virus in one-humped camels (Camelus dromedarius) population in northeast of Iran. J Vect Dis 51:62–65
Ergonul O (2006) Crimean-Congo haemorrahgic fever. Lancet Infect Dis 6:203–214
Chinikar S, Ghiasi SM, Hewson R, Moradi M, Haeri A (2010) Crimean-Congo hemorrhagic fever in Iran and neighboring countries. J Clin Virol 47:110–114
Saidi S, Casals J, Faghih MA (1975) Crimean hemorrhagic fever-Congo (CHF-C) virus antibodies in man and domestic and small mammals in Iran. Am J Trop Med Hyg 24:353–357
Mild M, Simon M, Albert J, Mirazimi A (2010) Towards an understanding of the migration of Crimean-Congo hemorrhagic fever virus. J Gen Virol 91:199–207
Chinikar S, Shah-Hosseini N, Bouzari S, Jalali T, Shokrgozar MA, Mostafavi E (2013) New circulating genomic variant of Crimean-Congo hemorrhagic fever virus in Iran. Arch Virol 158:1085–1088
Keshtkar-Jahromi M, Sajadi MM, Ansari H, Mardani M, Holakouie-Naieni K (2013) Crimean-Congo hemorrhagic fever in Iran. Antiviral Res 100:20–28
Sureau P, Klein JM, Casals J, Digoutte J, Salaun J, Piazak N (1980) Isolation of Thogoto, Wad medani, Wanowrie and Crimean-Congo hemorrhagic fever viruses from ticks of domestic animals in Iran. Ann Virol (Inst Pasteur) 131E:185–200
Chinikar S, Shayesteh M, Khakifirouz S, Jalali T, Rasi Varaie FS, Rafigh M, Mostafavi E, Shah-Hosseini N (2013) Nosocomial infection of Crimean-Congo haemorrhagic fever in eastern Iran: case report. Travel Med Infect Dis 11:252–255
Logan TM, Linthicum KJ, Bailey CL, Watts DM, Dohm DJ, Moulton JR (1990) Replication of Crimean-Congo hemorrhagic fever virus in four species of ixodid ticks (Acari) infected experimentally. J Med Entomol 27:537–542
Nabian S, Rahbari S, Changizi A (2009) The distribution of Hyalomma spp. ticks from domestic ruminants in Iran. Med Vet Entomol 23:281–283
Schwarz TF, Nsanze H, Longson M (1996) Polymerase chain reaction for diagnosis and identification of distinct variants of Crimean-Congo hemorrhagic fever virus in the United Arab Emirates. Am J Trop Med Hyg 55:190–196
Burt FJ, Leman PA, Smith JF, Swanepoel R (1998) The use of a reverse transcription-polymerase chain reaction for the detection of viral nucleic acid in the diagnosis of Crimean-Congo haemorrhagic fever. J Virol Methods 70(2):129–137
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Fakoorziba MR, Neghab M, Alipour H, Moemenbellah-Fard MD (2006) Tick-borne Crimean-Congo haemorrhagic fever in Fars province, southern Iran: Epidemiologic characteristics and vector surveillance. Pak J Biol Sci 9:2681–2684
Fakoorziba MR, Golmohammadi P, Moradzadeh R, Moemenbellah-Fard MD, Azizi K, Davari B, Alipour H, Ahmadnia S, Chinikar S (2012) Reverse transcription PCR-based detection of Crimean-Congo hemorrhagic fever virus isolated from ticks of domestic ruminants in Kurdistan province of Iran. Vect Zoonotic Dis 12:794–799
Moemenbellah-Fard MD, Benafshi O, Rafinejad J, Ashraf H (2009) Tick-borne relapsing fever in a new highland endemic focus of western Iran. Ann Trop Med Parasitol 103:529–537
Nuttall PAN (2001) Crimean-Congo haemorrhagic fever. In: Service MV (ed) Encyclopedia of arthropod-transmitted infections of man and domesticated animals. CABI Publishing, Wallingford, UK, pp 126–132
Hassanzadeh J, Mohammadbeigi A, Eshrati B, Moemenbellah-Fard MD (2012) Estimation of the regional burden of non-communicable diseases due to obesity and overweight in Markazi province, Iran, 2006-2007. J Cardiovasc Dis Res 3:26–31
Azizi K, Moemenbellah-Fard MD, Fakoorziba MR, Fekri S (2011) Gerbillus nanus (Rodentia: Muridae): a new reservoir host of Leishmania major. Ann Trop Med Parasitol 105:431–437
Azizi K, Moemenbellah-Fard MD, Kalantari M, Fakoorziba MR (2012) Molecular detection of Leishmania major kDNA from wild rodents in a new focus of zoonotic cutaneous leishmaniasis in an oriental region of Iran. Vect Zoonotic Dis 12:844–850
Moemenbellah-Fard MD, Saleh V, Banafshi O, Dabaghmanesh T (2012) Malaria elimination trend from a hypo-endemic unstable active focus in southern Iran: predisposing climatic factors. Pathog Glob Health 106:358–365
Sharifi-Mood B, Metanat M, Rakhshani F, Shakeri A (2011) Co-infection of malaria and Crimean-Congo hemorrhagic fever. Iran J Parasitol 6:113–115
Chinikar S, Hezareh Moghadam A, Parizadeh SMJ, Moradi M, Bayat N, Zeinali M, Mostafavi E (2012) Seroepidemiology of Crimean-Congo hemorrhagic fever in slaughter house workers in north eastern Iran. Iran J Publ Health 41:72–77
Mostafavi E, Haghdoost A, Khakifirouz S, Chinikar S (2013) Spatial analysis of Crimean-Congo hemorrhagic fever in Iran. Am J Trop Med Hyg 89(6):1135–1141
Tonbak S, Aktas M, Altay K, Azkur AK, Kalkan A, Bolat Y, Dumanli N, Ozdarendeli A (2006) Crimean-Congo hemorrhagic fever virus: Genetic analysis and tick survey in Turkey. J Clin Microbiol 44:4120–4124
Telmadarraiy Z, Moradi AR, Vatandoost H, Mostafavi E, Oshaghi MA, Zahirnia AH, Haeri A, Chinikar S (2008) Crimean-Congo hemorrhagic fever: A seroepidemiological and molecular survey in Bahar, Hamadan province of Iran. Asian J Anim Vet Adv 3:321–327
Tahmasebi F, Ghiasi SM, Mostafavi E, Moradi M, Piazak N, Mozafari A, Haeri A, Fooks AR, Chinikar S (2010) Molecular epidemiology of Crimean-Congo hemorrhagic fever virus genome isolated from ticks of Hamadan province of Iran. J Vect Dis 47(4):211–216
Gould EA, Higgs S (2009) Impact of climate change and other factors on emerging arbovirus diseases. Trans R Soc Trop Med Hyg 103:109–121
Pfäffle M, Littwin N, Muders SV, Petney TN (2013) The ecology of tick-borne diseases. Int J Parasitol 43:1059–1077
Mahzounieh M, Dincer E, Faraji A, Akin H, Akkutay AZ, Ozkul A (2012) Relationship between Crimean-Congo hemorrhagic fever virus strains circulating in Iran and Turkey: possibilities for transborder transmission. Vect Zoonotic Dis 12:782–785
Walker JG, Klein EY, Levin SA (2014) Disease at the wildlife-livestock interface: Acaricide use on domestic cattle does not prevent transmission of a tick-borne pathogen with multiple hosts. Vet Parasitol 199:206–214
Mostafavi E, Chinikar S, Bokaei S, Haghdoost A (2013) Temporal modeling of Crimean-Congo hemorrhagic fever in eastern Iran. Int J Infect Dis 17:e524–e528
Yesilbag K, Aydin L, Dincer E, Alpay G, Girisgin AO, Tuncer P, Ozkul A (2013) Tick survey and detection of Crimean-Congo hemorrhagic fever virus in tick species from a non-endemic area, South Marmara region, Turkey. Exp Appl Acarol 60(2):253–261
Christova I, Gladnishka T, Taseva E, Kalvatchev N, Tsergouli K, Papa A (2012) Seroprevalence of Crimean-Congo hemorrhagic fever virus, Bulgaria. J Med Virol 84(4):608–614
Olcay Hekimoglu O, Ozer N, Ergunay K, Ozkul A (2012) Species distribution and detection of Crimean Congo hemorrhagic fever virus (CCHFV) in field-collected ticks in Ankara Province, Central Anatolia, Turkey. Exp Appl Acarol 56(1):75–84
Tekin S, Bursali A, Mutluay N, Keskin A, Dundar E (2012) Crimean-Congo hemorrhagic fever virus in various ixodid tick species from a highly endemic area. Vet Parasitol 186(3–4):546–552
Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST (2006) Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol 80(17):8834–8842
Zeller HG, Cornet JP, Diop A, Camicas JL (1997) Crimean-Congo hemorrhagic fever in ticks (Acari: Ixodidae) and ruminants: field observations of an epizootic in Bandia, Senegal (1989-1992). J Med Entomol 34(5):511–516
Shepherd AJ, Swanepoel R, Cornel AJ, Mathee O (1989) Experimental studies on the replication and transmission of Crimean-Congo hemorrhagic fever virus in some African tick species. Am J Trop Med Hyg 40(3):326–331
Alam MM, Khurshid A, Sharif S, Shaukat S, Rana MS, Angez M, Zaidi SSZ (2013) Genetic analysis and epidemiology of Crimean Congo hemorrhagic fever viruses in Baluchistan province of Pakistan. BMC Infect Dis 13:201
Acknowledgments
The present paper was extracted from an approved MSc student thesis (No: 92-6700, dated 22 Sep 2013) conducted by the second author, Mr. A.A. Naddaf-Sani. Appreciation and thanks are due to the Vice-Chancellor for Research and Technology at SUMS for permitting the use of facilities at the university, Ms. T. Dabaghmanesh for help with office work, and Dr. T. Jalali, Ms. S. Khakifirouz, and Dr. N. Shah-Hosseini of the Pasteur Institute, Tehran, for collaborations on GenBank registrations, distance table and phylogenetic tree construction. This investigation was financially supported by an approved contract research plan (92-6700) awarded to MRF by Shiraz University of Medical Sciences (SUMS).
Conflict of interest
None declared.
Ethical approval
Not required.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fakoorziba, M.R., Naddaf-Sani, A.A., Moemenbellah-Fard, M.D. et al. First phylogenetic analysis of a Crimean-Congo hemorrhagic fever virus genome in naturally infected Rhipicephalus appendiculatus ticks (Acari: Ixodidae). Arch Virol 160, 1197–1209 (2015). https://doi.org/10.1007/s00705-015-2379-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2379-1