Abstract
A new species, Barleria lavaniana (Acanthaceae), is described and illustrated. It resembles B. longiflora but differs from it in the characters of calyx, stamen, style and stigma. Stem anatomy reveals that vessel elements are mostly angular with a short tail at one end and a long tail on the other end. Xylem rays are usually biseriate and xylem fibres relatively thin-walled. Interxylary phloem is generally characterized by a single companion cell. The somatic chromosome number was 2n = 2x = 40. The karyotype is moderately symmetrical and fell in Stebbins’ 3b category. Chromosomes have median and submedian region centromeres. Based on cpDNA data, the new species is placed in subgenus Barleria and shows phylogenetic affinity towards B. longiflora and B. acuminata. Our findings suggest the need for comprehensive phylogenetic analyses of the genus Barleria based on high-throughput sequencing data to understand species interrelationships and biogeography.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Barleria L. (Acanthaceae Juss.) was described on the basis of a specimen collected from India (Linnaeus 1753). It is a large, polymorphic, widespread genus of herbs, shrubs, rarely climbers or trees comprising some 300 species (Balkwill and Balkwill 1998; Darbyshire and Luke 2016). The maximum representation of the genus is seen in Africa with two centres of diversity, viz. tropical East Africa (ca. 80 species) and southern Africa (ca. 70 species) (Balkwill and Balkwill 1998). Recently, Darbyshire et al. (2019a) revised the tribe Barlerieae (Acanthaceae) in Angola and Namibia and recorded 32 species. Barleria can be easily distinguished from other genera of Acanthaceae by a combination of three characters, i.e. a four-partite calyx with two large outer segments and two smaller inner ones, globose honey-combed pollen grain and the predominance of double cystoliths in the epidermal cells (Balkwill and Balkwill 1998). Balkwill and Balkwill (1997), while classifying Barleria, subdivided it into two subgenera (subg. Barleria and Prionitis (Nees) C.B.Clarke) and seven sections (sect. Barleria, Chrysothrix M.Balkwill, Cavirostrata M.Balkwill, Fissimura M.Balkwill, Stellatohirta M.Balkwill, Somalia (Oliv.) Lindau and Prionitis (Nees) Lindau). Darbyshire et al. (2019b) provided a revised classification of Barleria wherein two subgenera, viz. Barleria and Prionitis, are recognized. Subgenus Barleria now includes species previously placed under sections Barleria, Chrysothrix and Fissimura. The species of section Cavirostrata are now included in section Somalia. Darbyshire et al. (2019b) recognized only three sections. These sections (Prionitis, Somalia and Stellatohirta) are placed under subgenus Prionitis.
The taxonomy of Barleria in India has been revised by Shendage and Yadav (2010). The genus now comprises 29 species, one subspecies and one variety (modified after Shendage and Yadav 2010). Species of the section Stellatohirta are not reported from India (Shendage and Yadav 2010). Subgenus Barleria has the largest representation with nineteen species. Section Somalia is represented by seven species, whereas Prionitis is the smallest section with three species (modified after Ravikumar et al. 2016). There are 17 endemic taxa (B. acuminata Wight ex Nees, B. courtallica Nees, B. cuspidata Heyne ex Nees, B. durairajii K.Ravik., D.Naras., Devanath. & Gnanasek., B. gibsonii Dalzell, B. grandiflora Dalzell, B. involucrata var. elata Nees, B. lawii T.Anderson, B. longiflora L.f., B. montana Nees, B. morrisiana E.Barnes and C.E.C.Fisch., B. pilosa B.Heyne ex Nees, B. prattensis Santapau, B. sepalosa C.B.Clarke, B. stocksii T.Anderson, B. terminalis (Nees) C.B.Clarke and B. tomentosa Roth.).
As a part of ongoing cytotaxonomic studies on Barleria (Gosavi et al. 2011; Joshi et al. 2016), we collected interesting specimens of Barleria from the Agashiv Hills, Karad taluka, Satara district, a southern part of the Indian state of Maharashtra in 2017. After critical scrutiny of the relevant literature and the examination of specimens as well as images of Barleria housed at different herbaria (B, BLAT, BSI, K, SUK and TBGT), it was determined that the specimens belong to a hitherto undescribed species of Barleria subgenus Barleria. The present paper describes and illustrates the new species. Karyotype and stem anatomy details of the new species and related species are also provided. Furthermore, a molecular phylogeny of the genus Barleria was constructed based on cpDNA data and a discussion on the phylogenetic placement of the new species is added.
Materials and methods
Taxonomy
The morphological analysis and description are based on observations made in the field and examination of live specimens. Identity of specimens was ascertained by consulting protologues, type specimens and specimens of related species. We studied the specimens housed at BLAT, BSI, CAL, CALI and SUK. Types and voucher specimens in the virtual herbaria (Global Plants on JSTOR and websites of BM, E, K and N) were viewed. The acronyms of herbaria consulted were obtained from Index Herbariorum (http://sweetgum.nybg.org/science/ih/). Descriptions were made following the terminology of Hickey and King (2001).
Cytogenetics
Mitotic studies were conducted on root tips obtained from germinated seeds. The procedure has been detailed elsewhere (Joshi et al. 2016). For karyotype analysis, ten well-separated somatic chromosome plates were selected. Karyotype asymmetry was determined using the categories of Stebbins (1971) and the CVCL (coefficient of variation of chromosome length) and MCA (mean centromeric asymmetry) as proposed by Peruzzi and Eroğlu (2013).
Stem anatomy
Samples from the main stems of Barleria lavaniana and B. longiflora were collected from five individuals growing naturally at Agashiv hills, Karad taluka, Satara district, Maharashtra and Vellar, Mettur Taluka, Salem district, Tamil Nadu, respectively. Stem pieces of 5–8 mm in diameter and 50–60 mm in length were collected 5 cm above the ground level for both the species and fixed in FAA (Berlyn and Miksche 1976). After 24 h of fixation, these samples were transferred to 70% ethanol for further processing and storage. Fixed samples were cut into 20–25-mm-long pieces, and 16–20-µm-thick sections were obtained in various plains, viz. transverse, tangential and radial longitudinal view, using a Leica SM2010R sliding microtome. Sections were stained with a safranin–astra blue combination (Srebotnik and Messener 1994), dehydrated through an ethanol–xylene series and mounted in dibutyl phthalate xylene (DPX). Micro-photographs were taken using a Leica DFC 295 fire wire digital camera attached to a Leica DM 2000 research microscope.
To study the morphology and dimensional details of the vessel elements and xylem fibres, small portions of the secondary xylem adjacent to the cambial ring were sliced into small pieces and macerated with Jeffrey’s fluid (Berlyn and Miksche 1976). After 24–36 h of treatment with Jeffrey’s fluid at 55–60 °C, treated wood pieces were washed thoroughly with distilled water and stained with 0.5% aqueous safranin. Lengths of the sieve tube elements were measured directly from the tangential longitudinal sections while widths were measured in transverse sections as suggested by the IAWA committee on bark terminology (Angyalossy et al. 2016). The tangential diameter of the vessel lumen was also obtained from the transverse section. Thirty random measurements for each cell type were used to obtain the mean and standard deviation. Wood description follows the IAWA committee (Wheeler et al. 1989) and Carlquist (2001).
Molecular phylogeny
Twenty taxa representing both the genus Barleria and the outgroup (Ruellia) species were sampled for phylogenetic analyses. Total genomic DNA was extracted from young and fresh leaves of Barleria longiflora, B. acuminata and the new species, B. lavaniana, using the modified CTAB method (Paterson et al. 1993) with some modifications. One gram of leaf tissue was placed in a mortar and pestle at − 20 °C for 10–15 min and then ground in the presence of 2 mL CTAB extraction buffer (Tris–HCl 100 mM, pH 8.0, 20 mM EDTA, 1.4 M NaCl and 2% CTAB) and incubated at 65 °C for 35 min. Chloroform–octanol (24:1) extraction was carried out to remove proteins. Iso-propanol was used to precipitate DNA. Two plastid regions, namely rbcL and matK, were amplified and sequenced as per Tamboli et al. (2018). DNA sequences were checked and edited by using Sequencher v 5.1 (Gene Codes Corporation 2012). Sequences of rbcL and matK regions were submitted to the GenBank NCBI database, and their accession numbers are shown in Table 1.
For phylogenetic analyses, we retrieved the rbcL and matK sequences of Barleria species from NCBI GenBank database. Multiple sequence alignment of all these sequences was carried out using MUSCLE (Edgar 2004) implemented in MEGA 7 (Kumar et al. 2016). The rbcL and matK sequence datasets were concatenated into a single matrix using BioEdit v. 7.2.6 (Hall 1999). The incongruence among these two sequence datasets was determined by using incongruence length test (ILD) (Farris et al. 1994). This test was performed on the concatenated dataset in PAUP 4.0a152 (Swofford 2002) with 100 homogeneity replicates, random taxon addition, holding 10 trees at each step tree-bisection-swapping (TBR) searches, MULTREES option in effect with maxtree set to 100. Phylogenetic analyses were carried out using three methods: maximum likelihood (ML), maximum parsimony (MP) and Bayesian analysis. Maximum likelihood (ML) and maximum parsimony (MP) were conducted in PAUP 4.0a152 (Swofford 2002), and Bayesian analysis was performed in MrBayes v. 3.0 (Ronquist and Huelsenbeck 2003). The best-fit nucleotide model for the concatenated dataset was selected using the automated model test implemented in PAUP 4.0a152 (Swofford 2002) based on Akaike information criterion (AIC). The best model shown was TVM + G. As this model is not present in PAUP 4.0a152 and MrBayes v. 3.0, we followed the next most complex model available in PAUP 4.0a152 and MrBayes v. 3.0 which is GTR + G. Maximum parsimony analyses were conducted in PAUP 4.0a152 (Swofford 2002) with 1000 bootstrap replicates. MP heuristic searches used 10 random taxa addition (holding 10 trees at each step), and tree-bisection-reconnection (TBR) was used as branch-swapping algorithm; ‘MulTrees’ options were set in effect. Maximum likelihood trees were drawn based on the best-fit nucleotide model with 1000 bootstrap replicates. For Bayesian phylogenetic analysis, the Markov chain Monte Carlo chains (MCMC) were run for 1 million generations. The convergence occurred when standard deviation (SD) of split frequencies fell below 0.01. The first 25% of MCMC generations were discarded as burn-in. Posterior probability values were used to estimate branch support.
Results
Stem anatomy
General structure of the secondary xylem: The secondary xylem of both the species is diffused porous with indistinct growth rings (Fig. 1a–d). It is composed of vessels, tracheids, fibres, axial and ray parenchyma cells while strands of interxylary phloem are distributed randomly (Fig. 1e, f). In both species, a single sieve element differentiates as an interxylary phloem derivative (Fig. 2e–f), but occasionally it can be in a group of 2–3 sieve elements. Vessels are mostly in radial multiples of two to several cells arranged in radial multiples (Fig. 2e, f) with alternate bordered pits on their lateral walls. Xylem rays are uni- to biseriate, heterocellular and vertically upright (Fig. 2c, d) and several cells in height. Axial parenchyma cells are thick-walled, extremely rare or nearly absent, difficult to distinguish in transverse view, scanty paratracheal and associated with vessel elements. In both species, xylem fibres are septate, possessed a distinct nucleus (Fig. 2c) and were oval to oblong or elliptic in outline (Fig. 2c).
a–f Structure of secondary xylem of Barleria lavaniana (a, c, e) and B. longiflora (b, d, f) in transverse view. a Gross structure of main stem of B. lavaniana showing cortex, secondary xylem and pith. Note the arrangement of vessels, b gross structure of main stem of B. longiflora. Note the number of vessels in radial multiples, c relatively enlarged view of a showing structure of secondary xylem and arrangement of vessels in radial multiples (arrowheads), d enlarged view of b showing arrangement of vessels. Arrowheads indicate radial multiples of vessels. Note that the lumen diameter of vessels and sieve tube elements is more as compared to B. lavaniana, e secondary xylem showing angular vessels (arrowhead) with relatively thin-walled xylem derivatives as compared to B. longiflora, f secondary xylem showing oval to circular vessels (arrow) and thick-walled xylem derivatives as compared to former species. Arrowheads showing group of 2–3 sieve elements. Scale bar a, b = 1 mm; c, d = 200 µm; e, f = 100 µm
Transverse (a, b, e, f) and tangential longitudinal (c, d) view of Barleria lavaniana (a, c, e) and B. longiflora (b, d, f). a Structure of secondary xylem. Note the angular outline of the vessels (arrowheads). Arrow indicates interxylary phloem, b oval to circular outline of the vessels (arrowheads). Arrow showing interxylary phloem, note the relatively wider sieve tube elements as compared to a, c uni-biseriate and heterocellular rays with vertically upright cells (arrows). Arrowheads showing nucleus in the fibre lumen while curved arrow shows septa in xylem fibre, d exclusively uniseriate rays composed of vertically upright and heterocellular rays (arrowheads). Arrow showing fusiform-shaped nucleus in one of the fibre, e interxylary sieve tube element showing relatively large associated parenchyma (arrow) and sieve tube element with single companion cell. Note the interxylary sieve tube element without associated axial parenchyma (arrowheads), f interxylary sieve tube elements with associated parenchyma (arrow). Note the adjacent sieve element without associated axial parenchyma. Arrowhead showing two companion cells with every sieve element, also note the larger diameter of sieve elements as compared to e. a–d: scale bar = 50 µm; e, f: scale bar = 20 µm
Barleria lavaniana is characterized by the presence of radial multiples of several vessels (4–21) in each radial file, while solitary vessels are rare. Vessels are mostly angular in outline (Figs. 1e, 2a), the perforation plate is simple with a long tail only on one end (Fig. 3a) in some elements, whereas very few of them show a tail on both the ends (Fig. 3a). Vessel elements are 584–631 (607 ± 7.289) µm in length and 40–59 (46.5 ± 2.046) µm in tangential diameter. Xylem rays are uni-biseriate, heterocellular with vertically upright cells (Fig. 2c). Rays are 227–316 (271 ± 3.935) µm in height and 10–29 (19.5 ± 4.087) µm in width. The length of the interxylary sieve tube elements ranges from 308 to 378 (342 ± 3.859) µm and is relatively smaller in diameter, i.e. 10–18 (13 ± 2.398) µm. In comparison with associated axial parenchyma, they are narrow in diameter and possess simple sieve plates with single companion cells (Fig. 2e). Associated axial parenchyma may be present or absent, and if present, the associated parenchyma has a larger diameter (Fig. 2e) than the sieve elements.
Structure and shape of vessel elements reconstructed from macerated material of Barleria lavaniana (a) and B. longiflora (b), a range of vessel morphology, structure and dimensions. Arrowheads showing tail at only one end. Arrows showing elongated tails on both the ends of vessel elements, b variation in vessel morphology. Note the elongated tail on both ends of the vessel elements (arrowheads). Figure 9a, b: scale bar = 100 µm
In contrast, vessels in B. longiflora are oval to circular (Figs. 1f, 2b) in outline (not angular) and in radial multiples of 2–6 or occasionally 8 vessels in each radial file (Fig. 1d, f). Vessel perforations are simple with a long tail on both end walls in most of the elements (Fig. 3b). In comparison with B. lavaniana, vessel elements in B. longiflora are shorter and wider (Table 2). Vessel walls are relatively thick compared to B. lavaniana and measured 5–8 (6 ± 0.738) µm thick. Xylem rays are exclusively uniseriate, heterocellular with vertically upright cells (Fig. 2d). Rays are 227–319 (278 ± 4.568) µm high and 10–19.8 (15.3 ± 3.752) µm wide. In comparison with B. lavaniana, interxylary sieve tube elements are shorter, measuring 298–338 (324.7 ± 2.834) µm while their diameter is greater, i.e. 21–29 (26 ± 2.229) µm. B. longiflora has simple sieve plates with mostly 2 companion cells (Fig. 2f) while single companion cell is observed occasionally. If present, associated axial parenchyma cells are smaller in diameter than the sieve elements (Fig. 2f).
Cytogenetics
Barleria lavaniana and B. longiflora have a diploid number of 2n = 40 (Fig. 4). The former has mean chromosome length of 2.78 ± 0.57 µm, whereas it is 3.22 ± 0.77 µm in the latter. Higher values of CVCL (33.25) and MCA (23.89) are observed for B. longiflora. Karyotype formulae for B. lavaniana and B. longiflora were 10 m + 10 sm and 6 m + 14 sm, respectively. The karyotype of both species is moderately symmetrical and fell in Stebbins’ 3b category (Fig. 5).
Molecular phylogeny
The aligned length of rbcL + matK data for the set of 20 taxa studied is 956 bp. Conserved sites, variable sites and parsimony informative sites for the combined data set were 868, 88 and 66, respectively. The results of ILD test between rbcL and matK dataset showed no significant incongruence between these datasets (P value = 0.03); hence, we combined both datasets (rbcL + matK) to construct the phylogenies.
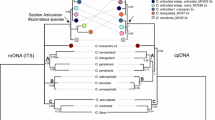
The tree with the highest log likelihood value (− 1933.370) is shown (Online Resource 1, Fig. 6a). In the case of the maximum parsimony analysis, the most parsimonious tree with length = 101 is shown for rbcL + matK data (Online Resource 1, Fig. 6b) with consistency index (CI) = 0.930 and retention index (RI) = 0.950. Bayesian phylogenetic analysis presented in Fig. 3 shows that the new species B. lavaniana has been placed in the clade representing subgenus Barleria and grouped with B. longiflora and B. acuminata with Bayesian PP value = 1, ML BS = 96 and MP BS = 95.
Discussion
Taxonomy
Barleria lavaniana belongs to the subgenus Barleria because it shares the following characters with members of this subgenus: capsule without a prominent beak, 4 seeds, stellate indumentum, etc. The species differs from B. longiflora in the characters listed in Table 3. Barleria lavaniana can be easily differentiated from most of the species (except B. acuminata and B. longiflora) of the subgenus in that it has stellate indumentum and a long corolla tube. Subgenus Barleria now comprises twenty species in India.
Stem anatomy
Histologically, B. lavaniana and B. longiflora showed considerable variation in vessel grouping, although both possessed vessels in radial multiples. In the case of B. lavaniana, the number of vessels in each radial file was higher (4–21), whereas in B. longiflora 2–6 vessels were frequent and up to 8 vessels occasionally were observed. Lens et al. (2009) hypothesized that vessel grouping pattern is a taxonomically important character in Apocynaceae. In the present study, vessel morphology, and vessel element length and diameter also exhibited significant difference between these species. However, vessel elements are sensitive to climatic conditions (ecological conditions), and so there is a possibility that these differences are just phenotypic. These modifications can be demonstrated by growing the same genetic stock at two or more localities (Bissing 1976; Akachuku and Burley 1979; Carlquist 2001). According to Stern and Greene (1958), qualitative features of vessels vary negligibly from one locality to another. However, the material studied here was growing under the same climatic conditions and was from the stems of same diameter. Vessel elements were angular and thin-walled in B. lavaniana and oval to circular with significantly thicker walls in B. longiflora. The presence of single companion cells in B. lavaniana (Fig. 2e) and two companion cells with each sieve element in B. longiflora (Fig. 2f) differentiates these species.
Cytogenetics
As Barleria has ornamental and medicinal significance (Joshi et al. 2016), considerable work on cytogenetics exists for the genus in India. The status of cytogenetics of the genus in India has been reviewed by Joshi et al. (2016). Presently, chromosome counts are not available for eight species (modified after Joshi et al. 2016). The diploid chromosome number in Barleria ranges from 2n = 24 (B. noctiflora) to 2n = 44 (B. grandiflora) with 2n = 40 being the most common diploid number indicating x = 10 as the base number for the genus (Joshi et al. 2016). B. lavaniana and B. longiflora both have 2n = 40 chromosomes, but their karyotypes differ in chromosome morphology. The former with equal number, i.e. 10 m and 10 sm chromosomes, had a more asymmetric karyotype than the latter where sm chromosomes dominate. Higher values of CVCL and MCA for B. longiflora further corroborate the advanced karyotype of the species. Both the species belong to subgenus Barleria; all the species of subgenus Barleria in India except B. morrisiana, B. nitida, B. pilosa and B. vestita have been investigated cytogenetically and possess 2n = 40 chromosomes.
Phylogenetic analyses
The genus Barleria has been limitedly studied from an evolutionary perspective. Baskaran (2016) studied phylogenetic relationships among some Barleria species based on only the rbcL region. In the present investigation, we analyse the phylogeny of Barleia species (mostly from subgenera, Barleria and Prionitis) based on cpDNA regions. Phylogenetic affinities of the new species (B. lavaniana) were also assessed. We follow the infrageneric classification provided by Darbyshire et al. (2019b) which is a modification of classification given by Balkwill and Balkwill (1997). The new species B. lavaniana is placed in the subgenus Barleria (Fig. 7) and shows phylogenetic affinity with B. longiflora and B. acuminata. Phylogenetic analyses in the present investigation had revealed that the species of subgenus Prionitis section Somalia (B. prattensis) formed a group with species of subgenus Barleria (B. cristata) within the clade representing subgenus Barleria with strong BI, ML and MP support (Fig. 7). Barleria ramulosa (subgenus Barleria) shows phylogenetic relationship with species of section Prionitis. Recently, Darbyshire et al. (2019b) provided the first comprehensive phylogenetic analyses of genus Barleria based on plastid intergenic spacers (trnS-G and ndhF-rpl32-trnL(UAG)) and nuclear ITS region. The results obtained in our study are at odds with the more in-depth analysis provided by Darbyshire et al. (2019a, b). This is due to the limited sampling of Barleria species and use of only two plastid regions (rbcL and matK) for construction of phylogeny. Consequently, there is low phylogenetic resolution and limited agreement with the morphological classification of the genus. Combined phylogenetic reconstruction (based on the plastid and nuclear region used by Darbyshire et al. 2019b) of Barleria sampling from Darbyshire et al. (2019b) and Barleria species analysed here can provide clear cut phylogenetic evidence for the new species as well as more agreement with the morphological classification. Darbyshire et al. (2019b) greatly contributed to our understanding of the evolution and complex pattern of morphological variation in Barleria. However, their phylogenetic results are unable to resolve relationships in the sampled taxa. These authors emphasized the need to add sampling from Indian subcontinent and Madagascar and also the use of high-throughput sequencing data to resolve the infrageneric relationship within the genus. This would also throw light on the biogeographic history of this pantropical genus.
Bayesian 50% majority rule consensus phylogenetic tree based on combined (rbcL + matK) data. Bayesian posterior probability, maximum likelihood and maximum parsimony bootstrap values (PP/ML/MP) are provided next to corresponding branches. The species sampled in this study are provided with their voucher specimen number in bracket after species name
Taxonomic treatment
Barleria lavaniana S.S.Patil, S.R.Yadav & Lekhak, sp. nov. —HOLOTYPE: India, Maharashtra, Satara district, Karad tehsil, Agashiv Hills, 723 m a. s. l., 17°14.087′N 74°09.216′E, 18 Sep 2018, S.S. Patil et al. 551 (holotype: CAL!; isotypes: BSI!, MH!, SUK!).
Etymology: The species is named in honour of Dr. Umesh Chandra Lavania, former Chief Scientist and Head, Division of Genetics and Plant Breeding, Central Institute of Medicinal and Aromatic Plants (CIMAP), Lucknow, India, and the Editor-in-Chief of The Nucleus, an International Journal of Cytology and Allied Topics, for his outstanding contribution in the field of plant cytogenetics.
Description: Perennial, profusely branched shrubs, up to 1.5 m high; branches woody at base, 1–1.2 cm in diameter, slender, bark off-white/light-brown, shallowly fissured, glabrous; young stems less woody, 0.3–0.6 cm in diameter, slender, tomentose; trichomes multicellular, dimorphic; stellate trichomes sessile, arms hyaline, more than ten, one extremely long, thrice the length of others, tapering towards apex, apex acute; glandular trichomes pilate, hyaline, tapering towards apex, tip hyaline initially, turning brownish-black at maturity. Leaves 3.6–13.7 × 2.5–8 cm, petiolate; lamina leathery, ovate; base rounded in young leaves, cuneate in old leaves (rarely oblique); apex acute; surface tomentose with stellate and glandular trichomes; adaxial surface light-green, density of trichomes less than abaxial surface; abaxial surface yellowish-green; venation camptodromous, one primary vein from base, 4–6 pairs of secondary veins arising along the length of the primary vein; petioles 0.5–6.6 cm long, light-green, tomentose. Inflorescence an axillary, scorpioid cyme, 2–3-flowered. Bracteoles two per flower, 0.6–1.6 × 0.2–0.4 cm, linear-lanceolate, persistent, light-green, tomentose, midrib conspicuous. Calyx of 4 dissimilar lobes, 2 outer, 2 inner, pale green, veins more than 10 from the base on the outer lobe, branching further to form a reticulate pattern, tomentose outside, pubescent within; outer adaxial lobe 2.2–3 × 0.6–1 cm, elliptic-lanceolate, margins entire, involute, apex acuminate; outer abaxial lobe 2.2–3 × 1–1.4 cm, lanceolate to ovate, margins entire, apex split into two teeth; inner (lateral) calyx lobes 1–1.7 × 0.2–0.3 cm, linear-lanceolate, apex acuminate, margins entire. Flowers 8–13.5 cm long, night blooming, corolla obscurely bilabiate, lobes subequal, pure white, puberulent outside, glabrous within; upper lip of 4 lobes, 2 adaxial lobes smaller than two lateral ones; adaxial lobes 2.2–2.4 × 0.8 × 1 cm, lanceolate-ovate, apex acute; lateral lobes 2.5–2.8 × 1.1–1.2 cm, lanceolate-ovate, apex acute; lower lip of 1 lobe, 2.5–2.7 × 0.9–1 cm, narrowly oblong, apex round to slightly retuse; tube cylindrical, 7–11 cm long, broad at base, gradually narrowing at apex, slightly inflated at the region of attachment of stamens, pale white, becoming pure white above the region of inflation, glandular pubescent, hairs more dense on the distal half of tube. Androecium comprising 2 stamens and 3 staminodes. Stamens 5.4–5.6 cm long, exserted, epipetalous; filaments pale white and flattened at base, white and slender above, glabrous except near base where glandular pubescent; anthers 0.8–0.9 cm long, white at anthesis, tinged bluish prior to dehiscence, brownish-black post-dehiscence; dehiscence introrse, longitudinal; thecae linear, muticous, parallel. Staminodes 1–1.2 cm long, glandular pubescent throughout; 2 staminodes with anther rudiments. Ovary, ovoid, two locular, glabrous except the region near the attachment of style; ovules 4; disc nectariferous, pale-yellow, cupular, enveloping the ovary for half of its length, hairy at apex, only on the margins; style 12.5–13.5 cm long, terete, white, glabrous except at the basal region (up to 2 cm from base towards apex); stigma 1–1.5 mm long, obliquely bilobed, hyaline. Capsule ellipsoid, 1.7–2.1 × 0.5–0.65 cm, stipitate, green when young, brown-black when mature, apex tapered into a beak, dehiscing explosively; upper half flattened and pubescent; lower half inflated and glabrous; nectary disc persistent. Seeds 4, 0.55–0.7 × 0.55–0.6 cm, covered with hygroscopic hairs. Figures 6 and 8.
Barleria lavaniana: a flowering twig, b flower with bracteoles and calyx, c corolla, opened along line between median abaxial lobe and lateral lobe (corolla split open), d staminodes (extended part of corolla tube removed), e gynoecium (pistil), f stamen, g abaxial outer calyx lobe, h adaxial outer calyx lobe, i, j inner calyx lobe, k bracteole, l unopened capsule, m seed
Diagnosis: Barleria lavaniana resembles B. longiflora. It differs in having outer calyx with truncate base usually ending in two apical teeth (vs. outer calyx cordate with entire apex in B. longiflora); the corolla lobes lanceolate-ovate (vs. corolla lobes oblong-obovoid in B. longiflora); the filament of the stamen being 5–5.2 cm long and presence of three staminodes (vs. the filament 1.7–1.8 cm long and presence of two staminodes in B. longiflora); the style being longer and pubescent at the base (vs. shorter style that is glabrous throughout in B. longiflora); the stigma 1–1.5 mm long obliquely 2-lobed (vs. stigma 0.5–0.7 mm long and 2-lobed in B. longiflora) and the capsule pubescent at apex (vs. the capsule glabrous throughout in B. longiflora) (Fig. 6 and Table 3).
Phenology: Flowering specimens were collected in September, whereas fruiting was observed in October and November.
Distribution: India.
Habitat: The species is known from Aurangabad, Osmanabad, Pune, and Satara districts of Maharashtra state, India (Fig. 9). It grows on rocky hill slopes above 600 m elevation.
Additional specimens examined: India, Maharashtra, Pune district, Kamalgi Ghat, 29 Oct 1905, W.A. Talbot 4441b, 2 Nov 1905, W.A. Talbot 4441c (BSI); Katraj Ghat, 5 Oct 1905, W.A. Talbot 4441a (BSI); Satara district, Agashiv hills, 16 Oct 2005, S.M. Shendage 2502 (SUK); Khambatki Ghat, 2 Nov 1964, R.S. Raghavan 104086 (BSI); 17 Nov 2005, S.M. Shendage 2514; Pateshwar, 9 Nov 2002, s.coll. 290 (SUK); Osmanabad district, Yedashi, 24 Sep 2017, S.S. Patil and P.V. Deshmukh 31 (SUK), 31 Oct 2017, S.S. Patil and P.V. Deshmukh 48 (SUK).
Conservation status: The species is known to exist in four locations. These localities were imported into GeoCAT (Bachman et al. 2011; http://geocat.kew.org/). The extent of occurrence (EOO) is estimated to be 31,106 km2 (far exceeding the 20,000 km2 upper limit for vulnerable status under the criterion B1), whereas its area of occupancy (AOO) is estimated to be 16 km2 (which falls within the limits for endangered status under the criterion B2). The habitat is constantly quarried for stones. The proposed category is endangered [EN B2ab(iii)] (IUCN 2001: version 3.1).
References
Akachuku AE, Burley J (1979) Variation of wood anatomy of Gmelina arborea Roxb. in Nigerian plantations. IAWA Bull 4:94–99
Angyalossy V, Pace MR, Evert RF, Marcati CR, Oskolski AA, Terrazas T, Kotina E, Lens F, Mazzoni-Viveiros SC, Angeles G, Machado SR, Crivellaro A, Rao KS, Junikka L, Nikolaeva N, Baas P (2016) IAWA list of microscopic bark features. IAWA J 37:517–615. https://doi.org/10.1163/22941932-20160151
Bachman S, Moat J, Hill A, de la Torre J, Scott B (2011) Supporting red list threat assessments with GeoCAT: geospatial conservation assessment tool. ZooKeys 150:117–126. https://doi.org/10.3897/zookeys.150.2109
Balkwill MJ, Balkwill K (1997) Delimitation and infra-generic classification of Barleria (Acanthaceae). Kew Bull 52:535–573. https://doi.org/10.2307/4110286
Balkwill MJ, Balkwill K (1998) Preliminary analysis of distribution patterns in a large, pantropical genus, Barleria L. (Acanthaceae). J Biogeogr 25:95–110. https://doi.org/10.1046/j.1365-2699.1998.251120.x
Baskaran K (2016) Analysis of phylogeny and evolutionary divergence of rbcL sequence of Barleria longiflora L.f. Int J Sci Res 5:544–548
Berlyn GP, Miksche JP (1976) Botanical microtechnique and cytochemistry. The Iowa State University Press, Ames
Bissing DR (1976) The effect of cultivation on the expression of anatomical features of the wood of selected dicotyledons. PhD Thesis, Claremont Graduate School, Claremont
Carlquist S (2001) Comparative wood anatomy systematic, ecological, and evolutionary aspects of dicotyledon wood. Springer, Berlin
Darbyshire I, Luke Q (2016) Barleria mirabilis (Acanthaceae): a remarkable new tree species from west Tanzania. Kew Bull 71:13. https://doi.org/10.1007/s12225-016-9622-0
Darbyshire I, Tripp EA, Chase FM (2019a) A taxonomic revision of Acanthaceae tribe Barlerieae in Angola and Namibia. Part 1. Kew Bull 74:5. https://doi.org/10.1007/s12225-018-9791-0
Darbyshire I, Fisher AE, Kiel CA, McDade LA (2019b) Phylogenetic relationships among species of Barleria (Acanthaceae, Lamiales): molecular data reveal complex patterns of morphological evolution and support a revised classification. Taxon 68:92–111. https://doi.org/10.1002/tax.12029
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113–131. https://doi.org/10.1186/1471-2105-5-113
Farris JD, Kallersjo M, Kluge AG, Bult C (1994) Testing significance of incongruence. Cladistics 10:315–319. https://doi.org/10.1111/j.1096-0031.1994.tb00181.x
Gene Codes Corporation (2012) Sequencher 5.1 gene codes corporation. Ann Arbor, Michigan. Available at: http://genecodes.com/
Gosavi KVC, Lekhak MM, Chandore AN, Yadav SR (2011) Karyology of Barleria grandiflora Dalzell (Acanthaceae), a potential ornamental endemic to Northern-Western Ghats of India. Nucleus 54:133–136. https://doi.org/10.1007/s13237-011-0040-2
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hickey M, King C (2001) The Cambridge illustrated glossary of botanical terms. Cambridge University Press, Cambridge
Wheeler EA, Baas P, Gasson P (eds) (1989) IAWA list of microscopic features for hardwood identification. IAWA Bull 10:219–332
IUCN (2001) IUCN Red List Categories and Criteria: Version 3.1. IUCN Species Survival Commission, IUCN, Gland and Cambridge
Joshi HS, Yadav PB, Lekhak MM, Yadav SR (2016) Cytogenetics of two endemic Barleria species (Acanthaceae) from the northern Western Ghats (India). Caryologia 69:170–174. https://doi.org/10.1080/00087114.2016.1152111
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molec Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lens F, Endress ME, Baas P, Jansen S, Erik Smets E (2009) Vessel grouping patterns in subfamilies Apocynoideae and Periplocoideae confirm phylogenetic value of wood structure within Apocynaceae. Amer J Bot 96(12):2168–2183. https://doi.org/10.3732/ajb.0900116
Linnaeus C (1753) Species Plantarum 2: 536–537
Paterson A, Brubaker C, Wendel J (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Pl Molec Biol Rep 11:122–127. https://doi.org/10.1007/BF02670470
Peruzzi L, Eroğlu H (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet 7:1–9. https://doi.org/10.3897/CompCytogen.v7i1.4431
Ravikumar K, Narasimhan D, Devanathan K, Gnanasekaran G (2016) Barleria durairajii (Acantha ceae): a new species from Tamil Nadu, India. Rheedea 26:136–141
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Shendage SM, Yadav SR (2010) Revision of the genus Barleria (Acanthaceae) in India. Rheedea 20:81–130
Srebotnik E, Messener K (1994) A simple method that uses differential staining and light microscopy to assess the selectivity of wood delignification by white rot fungi. Appl Environm Microbiol 60:1383–1386
Stebbins G (1971) Chromosomal evolution in higher plants. Edward Arnold, London
Stern WL, Greene S (1958) Some aspects of variation in wood. Trop Woods 108:65–71
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). version 4.0b10. Sinauer Associates, Sunderland. https://doi.org/10.1111/j.0014-3820.2002.tb00191.x
Tamboli AS, Yadav PB, Gothe AA, Yadav SR, Govindwar SP (2018) Molecular phylogeny and genetic diversity of genus Capparis (Capparaceae) based on plastid DNA sequences and ISSR markers. Pl Syst Evol 304:205–217. https://doi.org/10.1007/s00606-017-1466-z
Acknowledgements
Authors thank the Head, Department of Botany, Shivaji University, for providing necessary facilities. We are grateful to Dr. Kishore S. Rajput, Associate Professor, Department of Botany, The Maharaja Sayajirao University of Baroda, Vadodra, Gujarat 390002, for helping us with stem anatomy studies. SSP thanks Ministry of Environment, Forest and Climate Change, Govt. of India, for providing Junior Research Fellowship vide sanction letter F.No.10/04/2012-CS/BG dated 19/11/2013. AST thanks Council of Scientific and Industrial Research, New Delhi, for Senior Research Fellowship (SRF) (Award Number–09/816(0042)/2018-EMR-I and ACK No–113307/2K17/1). We are grateful to Dr. Shrikant P. Sutar, Department of Botany, Nowrosjee Wadia College, Maharashtra for the illustrations. We thank the reviewers for the critical comments that improved the content of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Ricarda Riina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendices
Appendix 1
Voucher information of Barleria longiflora.
India, s.d., s.coll., s.n. (E00892701, E00892710, E00892711) (E); s.d., J.G. Konig s.n. (10004760, 10004762, 10004763) (C); s.d., K. Narayana Iyer s.n. (Acc. No. 01899) (TBGT); s.d., W. Roxburgh s.n. (E00874943, E00892702); s.d., P. Russell s.n. (E00874942); 1961, Wight s.n. (E00892703, E00892704, E00892712) (E); 1961, Wight 2501c (E00892708) (E). Andhra Pradesh, Guntur district, Mellavagu, 3 Sep 1956, S.K. Wagh 3619, 3620, 3622 (BLAT), Nagarjunanagar, 13 Dec 1959, Sebastine 9751, Nagarjunakonda valley, 27 Nov 1961, K. Thothathri 9807 (CAL); Krishna district, Kondahalli, Jan 1887, J.S. Gamble 18576; Kundapalli, 25 Aug 1956, S.K. Wagh 3290, 3291 (BLAT); Kurnool district, Rangapurum R. F., 11 Oct 1983, J.S. Gamble 1706; Prakasam District, Gangavaram hill, 3 Dec 1983, R.K. Mohan 0319; Orvakal mountains, 8 May 2004, S.K. Nageervddin 622 (CAL). Karnataka, Raichur district, Bankaldoddi R.F., Deodurg, 14 Nov 1975, N.P. Singh 141692 (BSI). Kerala, Palakkad district, Attappadi, 12 Dec 1973, J.G. Menon 103 (TBGT). Tamil Nadu, Dharmapuri district, Dharmapuri, 7 Dec 1978, K.M. Mathew 19714 (CAL); Dindigul district, Sirumalai hills, 26 May 1958, J. Pallithanam 3462, 5 Feb 1959, J. Pallithanam 4288 (BLAT); Kanchipuram district, Pallavaram, s.d., s.coll., s.n. (E00892705) (E); Thirukazhukundram, s.d., K. Narayana Iyer s.n. (Acc. No. 01552) (TBGT); Vandalur hills, 10 Jan 1958, H. Santapau 22344 (BLAT); Kanayakumari district, Gomuki-Vellimalai, 7 Feb 1983, K. Ramamurthy 77365 (CAL); Nagapattinam district, Nagapattinam, s.d., Wight s.n. (E00892714) (E); Vandalur, 10 Jan 1958, K. Subramanyam 5413; Vandalur R. F., 3 Feb 1976, A.N. Henry 47153; North Arcot district, Sathanur dam forest, 26 Nov 1971, Vajravelu 52091 (CAL); Puducherry district, Pondicherry, 1834, M. Ferrollet s.n. (E00892706) (E); South Arcot district, Ginjee R.F., 17 Mar 1961, Sebastine 12171; Ginjee to Chokuptha, 18 Feb 1889, K. Ramamurthy 60308; Gomukhi dam, Thagari forest north, 19 Jan 1978, Ramamurthy 52852; 14 Dec 1978, Matthew and Perumal 20215 (CAL); Travancore, 1817, s.coll., 2501a (K001116222), 2501b (K001116224), 2501c (K001116223) (K); 1817, s.coll., 2501a (E00892707) (E); Tiruchirapalli district, Kolli Hills, Puliancholai, 30 Nov 1975, Anobaran 19570; Tirunelveli District, Kalkad hill, 7 Feb, C.E.C. Fischer 3873 (CAL).
Information on Electronic Supplementary material
Online Resource 1. Phylogenetic analyses using maximum likelihood and maximum parsimony method.
Online Resource 2. Aligned matrix of rbcL + matK dataset.
Rights and permissions
About this article
Cite this article
Patil, S.S., Tamboli, A.S., Yadav, S.R. et al. A new species of Barleria (Acanthaceae), its morphotaxonomy, cytogenetics and phylogenetic placement. Plant Syst Evol 305, 933–947 (2019). https://doi.org/10.1007/s00606-019-01613-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-019-01613-2