Abstract
An axenic culture of a cyanobacterium (strain HPS) was raised from a field specimen of greenish planktonic floccose mass from a local lake, Ganga Sagar. Intense morphological examination and comparative morphological assessment with the genera Fischerella and Hapalosiphon and all the known strains of the genus Westiellopsis indicated that the strain HPS differed in morphology with the closely related strains in trichome arrangement, size of vegetative cells, heterocytes, monocytes, and number of rows of main filaments. There were also differences in the habitat preference, being aquatic rather than terrestrial or subaerial. Intense ecological characterization of the habitat was performed through measurements of important physicochemical characteristics. 16S rRNA gene-based identification and phylogenetic placement indicated conclusively that the strain HPS was different from the most closely related strain Westiellopsis prolifica SAG 16.93. Phylogenetic inferences drawn in between all the branched heterocytous forms and subsequent 16S-23S ITS analyses and folding of the secondary structures revealed an entirely new form that is unknown till now. Subsequent nifD and rbcL gene-based phylogenetic assessments indicated that strain HPS is phylogenetically different from all the other previously known species of true branching cyanobacteria, along with also pointing toward the huge database inconsistencies in case of true branched cyanobacteria. Assessment of morphological and ecological differences along with comprehensive phylogenetic evaluation indicated that the strain HPS is a new species of the genus Westiellopsis and the name being proposed is Westiellopsis ramosa sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the bacteriological classification (Rippka et al. 1979; Anagnostidis and Komárek 1990; Castenholz 2001), true branching heterocytous cyanobacteria belonging to Subsection V (order Stigonematales) differ from Subsection IV (order Nostocales) on the basis of the plane of cell division. Longitudinal and oblique divisions bring about creeping branching out of transversely dividing main trichome. The true branching cyanobacteria display considerable morphological variation in shapes and dimensions of the cells, number of layers of cells and branching types. Gugger and Hoffmann (2004) classified the Stigonematales into a clade of four clusters separating on the basis of Y- and T-branching. Hoffmann et al. (2005) regrouped the orders Nostocales and Stigonematales into one subclass Nostochopycideae, as they were found to be monophyletic. Komárek et al. (2014) revised the taxonomic position applying a polyphasic approach, combining 16S rRNA data with morphological characteristics, including ultrastructure and ecological backdrop. The study reaffirmed that all heterocytous forms group in one order Nostocales, which was finally divided in eight families that included Symphyonemataceae (true branching “Y”), Hapalosiphonaceae (true branching “T”) and Stigonemataceae (true branching, multiseriate). The family Hapalosiphonaceae consists of genera like Fischerella Gomont ex Bornet & Flahault, Westiellopsis Janet, Nostochopsis Wood ex Bornet & Flahault, Hapalosiphon Nägeli in Kütz ex Bornet & Flahault, Mastigocladus Cohn ex Kirchner and Mastigocoleus Lagerheim ex Bornet & Flahault. These genera are closely related as per phylogenetic assessments as they form a monophyletic clade, but are intriguing and confusing on morphological scales (Gugger and Hoffmann 2004; Komárek and Mareš 2012; Dagan et al. 2013). Due to the confusing morphologies only, it is well established now that this family is in urgent need of polyphasic revisions (Jeeji-Bai 1972; Kaštovský and Johansen 2008).
The genus Westiellopsis is characterized by true branched filaments in which the main filaments may be uni- or biseriate (rarely multiseriate) and monoseriate T-shaped branches which are always thinner than the main filaments. It must be noted that all the described species of this genus have been reported to have almost spherical-rounded chroococcoid stages. The type species of this genus is Westiellopsis prolifica Janet (Janet 1941). Till date, apart from Westiellopsis prolifica, only three more species have been described viz., Westiellopsis indica Bourrelly (Bourrelly 1970), Westiellopsis interrupta Kanthamma (Jeeji-Bai 1972), and Westiellopsis iyengari Jeeji-Bai (Jeeji-Bai 1972).
Since the true branching heterocytous cyanobacteria are the most complex forms of cyanobacteria and believed to be the putative endosymbionts for photosynthetic life in eukaryotes (Dagan et al. 2013), it would be worthwhile to include more isolates for better understanding of evolutionary and phylogenetic lineages. In this communication, we describe a new species of the genus Westiellopsis according to the International Code of Nomenclature of Algae, Fungi, and Plants using morphological, ecological, and molecular traits.
Materials and methods
Sampling and culturing
The strain HPS was isolated from the sample collected from Ganga Sagar Lake, Jabalpur, India (23°10′N, 79°56′E). Water parameters were analyzed as follows (APHA 2012): BOD, 5-Day BOD Test; COD, Closed Reflux, Titrimetric Method; dissolved O2 (DO), Azide Modification; phosphate, Stannous Chloride Method; chloride, Argentometric Method; free CO2, Titrimetric Method; total and calcium hardness, EDTA Titrimetric Method and total alkalinity, Titration Method. Ammonia was determined by the Nesslerization method (Solarzano 1969). The collected natural sample was immediately analyzed microscopically to get an idea about the naturally occurring inherent microflora along with having a look at the morphology of the cyanobacteria present. Thereafter, pure colonies were raised on sterilized petri plates by the spread plate technique on 1.2% agar-solidified BG-11o medium (Rippka et al. 1979). After 20 days of growth, one or two colonies were picked up, washed thrice with deionized water and transferred to 1 mL of fresh liquid BG-11o medium. As the cultures started to disperse in the medium in 10–12 days, intact new filaments, viewed under 40X objective, were collected using sterilized 20-μL micro-capillaries (Sigma Aldrich) and dispensed into new batches of 1-mL medium. The step was repeated until purity of the culture was established by phase-contrast and bright-field microscopy, and by plating culture aliquots on Luria–Bertani medium. This novel microscopic dilution method was adopted to establish axenicity of the culture. The heterocytous cyanobacterial strain was then grown in 200 mL batches of the above medium contained in cotton stoppered Erlenmeyer flasks (capacity 500 mL). pH of the medium was adjusted to 7.2, and the culture was maintained in culture room under illumination of approximately 50–55 µ Em−2 s−1 with a photoperiod of 14/10 h light/dark cycle at 28 ± 2 °C. The culture was shaken twice a day.
Phenotypic analysis
The strain HPS was observed under Nikon YS100 microscope (Nikon, Japan). Microphotographs of the cyanobacteria were taken on Olympus BX53 (Olympus Corporation, Japan) fitted with ProgRes C5 camera (Jenoptik, USA). The shape of apical cells, shape, size, orientation, and other miscellaneous features of the filaments, vegetative cells, and heterocytes were visualized and observed with 50–100 measurements, recorded for each parameter. Structural details of the main filaments, such as the arising branches, pattern of branching, and presence/absence of hormocytes and plane of division were also observed. Measurements at 40× and 100× magnifications were taken for better resolution. The initial identification of the cyanobacteria was done using the keys of Desikachary (1959). Keys of Komárek (2013) were consulted for the final conclusion about the morphological description of the strain HPS.
Genomic DNA extraction and PCR
The DNA was isolated from 12- to 14-day-old log-phase culture using the Himedia Ultrasensitive Spin Purification Kit (MB505) with some modifications being done in the lysis step in which incubation time was increased for both the lysis solutions AL and C1. Amplification of the 16S rRNA gene was performed using the primers pA(5′-AGAGTTTGATCCTGGCTCAG-3′) and B23S (5′-CTTCGCCTCTGTGTGCCTAGGT-3′) (Edwards et al. 1989; Gkelis et al. 2005). The amplification of rbcL gene was done according to Singh et al. (2015a), while the nifD gene was amplified as per Singh et al. (2015b).
Phylogenetic analysis
Sequencing of the amplified products was carried out by the Sanger method using 96 capillaries and a 3730xl automated sequencer (Applied BioSystems). In case of the 16S rRNA gene and ITS region, a clean 1562-bp sequence was generated and the similarity search was piloted using the ‘Identify’ option of the Eztaxon database (http://www.eztaxon.org, Kim et al. 2012) with only validated cyanobacterial strains. Appropriate sequences from NCBI were also selected for the phylogenetic analysis using the 16S rRNA gene. For the rbcL and nifD genes, the sequences obtained were analyzed using the NCBI Web services with the BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi) tool and submitted to the NCBI database using the tool BankIt. The sequences were annotated for the coding regions using the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/).
Multiple sequence alignment was performed using CLUSTAL X, version 1.81 (Thompson et al. 1997). With an aim to obtain unambiguous data, the alignment was manually edited using DAMBE (Xia 2013). All the phylogenetic analyses were performed using the software package MEGA version-5 (Tamura et al. 2011). Analyses for the 16S rRNA gene phylogeny were performed using the validly published sequences from the Eztaxon database (Kim et al. 2012) along with selecting good quality sequences from NCBI. For the 16S rRNA gene based phylogeny of all the heterocytous forms, the suitable model of phylogeny with the lowest Bayesian Information Criterion (BIC) score was selected using the MEGA 5 program which led to the selection of the Kimura 2-parameter model (K2 + G + I) model having the lowest BIC score of 6350.038. For the 16S rRNA gene phylogeny of only the true branched forms, the K2 + G model was used. In case of the rbcL gene, the evolutionary history was inferred by using the maximum likelihood method based on Kimura 2-parameter model (K2 + G) model with the BIC score being 9877.951. The nifD gene was phylogenetically assessed through the K2 + G model with the BIC being 1883.642. It was assumed that the models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. For each model, the Akaike information criterion-corrected value (AICc), maximum likelihood value (lnL), and the number of parameters (including branch lengths) were also determined. The non-uniformity of evolutionary rates among sites was adjusted to be modeled by using a discrete gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (+I). Bootstrap values were deduced based on 1000 replications (Felsenstein 1985). In order to evaluate the robustness of the tree topology, clustering was performed using three different methods: neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) (Felsenstein 1981, 1985; Fitch 1971; Saitou and Nei 1987).
16S-23S ITS secondary structure analysis
Secondary structures of the 16S–23S internal transcribed spacer (ITS) region were determined carefully for two closely related taxa Westiellopsis sp. Ar73 (DQ786168) and Fischerella muscicola SAG 2027 (AM709634) present in the phylogenetic interpretations and further used for comparisons with the secondary structure of the strain HPS. These structures were transcribed and folded for all the relevant strains using the Mfold Web server (Zuker 2003).
Results
Habitat analysis
The various physicochemical characteristics of the water sample from the lake were measured so as to gain information about the general ecology of the strain HPS. The Ganga Sagar Lake (depth ~58 m) is an artificial lake with intimate interaction with neighboring cohorts of human population. The metabolic status is eutrophic to dystrophic. At the time of sampling, the water temperature was measured at 21 °C with the pH being 7.1. Crucial water quality parameters like BOD, COD, and DO were measured at 18, 28, and 0.28 ppm, respectively. Concentration of phosphate and inorganic nitrogen in the form of ammonia was evaluated at 0.3 and 4 mg/L, respectively, while the chloride concentration stood at 80 ppm. Free carbon dioxide amounted to 28 ppm, while total hardness of water was calculated at 150 ppm. Finally, the total alkalinity was determined to be 135 ppm, while total calcium was found to be 70 ppm.
Morphological evaluation
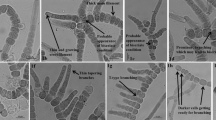
The various morphological characteristics of the strain HPS were studied carefully, and comparative assessment was made with the four Westiellopsis taxa known so far, i.e., W. prolifica, W. indica, W. interrupta, and W. iyengari (Table 1). The morphological features assessed with priority were the type of filament orientation, sheath and its overall distribution, visibility across the trichome, the shape and size of the vegetative cells, heterocytes, and ‘T’ branching pattern (Fig. 1). As a confirmatory cross-check, comparisons were also made with the genera Fischerella and Hapalosiphon. Uniseriate trichome, constricted cells in main trichome while cylindrical cells in branches, singly releasing monocytes from branch terminals, thinner branches relative to main trichome with occasional tapering are some of the diacritic features of strain HPS not found in Hapalosiphon. The strain HPS differed from the genus Fischerella also, in having terminal heterocytes (intercalary or lateral heterocytes are more common in Fischerella) and the absence of special superficial structures in cell wall (these special superficial structures are seen in Fischerella).
a Terminal cylindrical heterocyte. b Intercalary heterocyte; constricted cells of main axis. c Branches developing adjacent to each other but all in the same direction. d Intense branching with terminal heterocytes at both ends. e Constricted cells of branches; Constricted cells of main axis; Intense branching. f Differently shaped cells of main axis; Constricted cells of main axis. g Differently shaped heterocytes. h Intensely branched unidirectional filament. i Intense and rare bidirectional branching. j Initial development of branches. k Solitary, spherical monocytes emerging from branch endings; Intensely branched unidirectional filaments. Bar, 5 µm
Molecular evaluation
In the similarity search using only validated 16S rRNA sequences from the EzTaxon database, the strain HPS showed 97.40% sequence similarity with W. prolifica SAG 16.93 (AJ544086), 97.41% similarity with H. delicatulus IAM M-266 (AB093484) and 97.57% similarity with H. welwitschii (AY034793). Alternatively, the similarity search using all the sequences from NCBI yielded 97% similarity with Westiellopsis sp. Ar73 (DQ786168) and Fischerella muscicola SAG 2027 (AM709634).
The nifD gene sequence similarity was found to be closest with Fischerella sp. UTEX 1903 (AY196955), the pair-wise similarity being 94%. In case of the rbcL gene, the closest similarity was found to be 99% with F. muscicola UTEX 1829 (AB075910), while only 97 and 94% pair-wise similarity was found with H. hibernicus BZ-3-1(EU151924) and F. muscicola PCC7414 (AB075912), respectively (Table 2).
Phylogenetic analysis
In the total heterocytous cyanobacteria tree, the strain HPS was found to be clustered close to the strain W. prolifica SAG 16.93 (AJ544086) (Fig. 2; Online Resource 1) with the separating node having sound bootstrap support in case of the NL, ML, and MP algorithms. The strain clustered with a distinct and separate node with the other closely related strains H. welwitschii (AY034793), H. delicatulus IAM M- 266 (AB093484), N. lobatus 92.1 (AJ544080), and H. hibernicus BZ-3-1 (EU151900). The NJ and the MP trees also exhibited similar tree topologies with good bootstrap support at all the pertinent nodes. In the phylogenetic tree having all the branched forms (Fig. 3), strain HPS paired with Westiellopsis sp. NQAIF324 (KJ636972), though the bootstrap support was strong only in case of the NJ tree. Noticeably, the overall bootstrap support for the complete cluster having the strain HPS was strong (100/100/99) in case of all the trees, though the support was not significant at the internal nodes. Also, the complete cluster comprised of only two genera: Westiellopsis and Fischerella. In the rbcL gene-based ML/NJ/MP combined dendrogram (Online Resource 2–3), the strain HPS was again found to be separated from its closest relative F. muscicola UTEX 1829 (AB075910) with very good bootstrap support. In the ML tree, the strain HPS was clearly placed on a separate node in comparison to the closely related strain H. hibernicus BZ-3-1(EU151924) with very good bootstrap support. The NJ and the MP trees bootstrap values again exhibited similar pattern of clustering like the ML tree in case of the rbcL gene also. In the nifD gene-based ML/NJ/MP combined dendrogram (Online Resource 4–5), the pattern of clustering was again distinctive with the strain HPS, showing clear-cut separate clustering with appropriate phylogenetic distance with various species of the genus Fischerella. Also, the strain HPS was found to be separated from its closest relative member Fischerella sp. UTEX 1903 (AY196955) with good bootstrap support.
16S-23S ITS secondary structure analysis
The D1–D1′ helix was sequenced and it was found to have a length of 77 nucleotides. The folding of the D1–D1′ helix of the secondary structure of the strain HPS along with the two closely related species Westiellopsis sp. Ar73 (DQ786168) and Fischerella muscicola SAG 2027 (AM709634) showed differences in the folding patterns of HPS as compared to the other two strains as there were prominent differences in the base of the stem, the loop just above the stem and the topmost loop of the helix (Fig. 4). For the D1–D1′ helix, the base of the stem of HPS comprised of 6 bp’s which was unusual as the rest of the closely related species had just 4 bp stem. More perplexing was the presence of two unpaired bp’s in this region. Apart from the differences in the starting stem, the first loop above the base of the stem consisted of 2 uracils at the 3′ end instead of the single uracil being present in the other two closely related species. Also, the topmost loop consisted of 10 bp’s as compared to the other two having only 8 bp’s.
Discussion
We used morphological, ecological and molecular methods to characterize and identify a true branched heterocytous cyanobacterium that was isolated from a floating cyanobacterial mat from the corners of a freshwater lake from Jabalpur, India. It must be noted that the initial work on this culture started considering it as a member of the genus Hapalosiphon. Finally, in-depth studies confirmed it as a member of the genus Westiellopsis. It is well known that members of the family Hapalosiphonaceae form a monophyletic clade but due to their high morphological variability, it has been advocated that there are needs for polyphasic revisions (Gugger and Hoffmann 2004; Komárek and Mareš 2012; Dagan et al. 2013; Jeeji-Bai 1972; Kaštovský and Johansen 2008). The term polyphasic and what all constitutes a polyphasic study in cyanobacteria has been summarized aptly by Komárek (2016) where it is mentioned that the use of a combination of methods that focus on the morphological, ecological and genetic parameters ideally constitutes the polyphasic approach. It is further recommended that while genetic assessment will be the primary criterion, the morphological, ecophysiological, and ecological characteristics are also equally important secondary criteria that must be followed and which must also be selected carefully depending on the nature of sample.
Due to the complicated morphology of the true branched forms, we performed the morphological comparison of our strain with the genus Hapalosiphon. This comparative morphological assessment was done also taking into consideration the descriptions of Hapalosiphon in Desikachary (1959) and Komárek (2013). After clearing our doubts over Hapalosiphon, the morphology of the strain was compared with the other known true branched heterocytous taxa in Desikachary (1959) and Komárek (2013), and the strain was confirmed to be belonging to the genus Westiellopsis. On further investigations, it was found that till date only four species of this genus have been reported and all are mostly from the Indian subcontinent. Comparative assessments with the morphology of all the known strains provided a clear proof that the strain HPS differed from all the previously known species of Westiellopsis (Table 1). Although morphologically our isolate was very close to W. prolifica, yet certain properties of HPS, such as (a) main filaments—neither torulose nor flexuous, (b) cell/monocyte/heterocyte size—smaller, (c) number of rows—uniseriate and (d) akinetes—absent, are unique which distinguish it expressively from all the known species of Westiellopsis.
Nevertheless, we also assessed carefully the ecology and habitat characteristics of the strain HPS with the earlier reported Westiellopsis species and found clear evidence suggesting that while HPS was isolated from a floating mat of a freshwater lake, the others were soil isolates or collected from rice fields.
A point that needs mention when describing a new species of the genus Westiellopsis is that the chances of finding representative isolates of any of the four species of the genus Westiellopsis recorded from India is a difficult task, and we failed to find any of the strains from any known depository either from India or from any part of the world. More surprisingly, expect a few sequences of Westiellopsis prolifica, there was no sequence for any gene for the other three known taxa Westiellopsis indica (Bourrelly 1970), Westiellopsis interrupta (Jeeji-Bai 1972), and Westiellopsis iyengari (Jeeji-Bai 1972). This finding gains importance in view of the fact that in cases like these, authors need to systematically study the genus under consideration in many directions which sometimes may be difficult to decide. An important message of this work is that in difficult cases like these, there must be a pertinent literature survey before deciding the final taxonomic identity of a strain. After a thorough evaluation of all the available literature, our own data, morphology and ecology, the strain HPS certainly appeared as an interesting strain that needed to be assessed through genetic methods which essentially constitute the primary criterion of the polyphasic approach (Komárek 2016).
The molecular evaluation was based on the use of the 16S rRNA gene along with the rbcL and nifD gene-based phylogenetic studies. The 1562-bp 16S rRNA gene and ITS region-based phylogenetic studies showed the phylogenetic placement of the strain HPS with W. prolifica SAG 16.93, though the distance indicated that actually the strain HPS was different from W. prolifica SAG 16.93. Subsequent NJ and MP trees constructed also showed similar topology thus strengthening our phylogenetic observations. With a 97.40% similarity with W. prolifica SAG 16.93, the strain HPS also satisfied one of the primary criterion for species identity as laid by Strackerbrandt and Ebers (2006) when the threshold limit for species identity was raised to 98.7–99%. It must be noted that these rigid values of percentage identity do not really justify the enormous diversity that the microbial world offers; hence caution must be maintained in these types of studies where new taxa are being added. In order to decide the taxonomic affiliation of the strain HPS with a greater degree of confidence, we analyzed the phylogeny of many other branched heterocytous forms on the basis of the 16S rRNA gene. For this, we retrieved sequence-based information from previous works (Gugger and Hoffmann 2004; Tomitani 2004; Finsinger et al. 2008). Phylogenetic reconstructions using all the available pertinent 16S rRNA sequences indicated decisively toward few things which need mention. The phylogenetic affiliation of strain HPS was still with the genus Westiellopsis though the exact placement again indicated that it was different from any other isolate of Westiellopsis that was sequenced in the past. The tree also indicated towards the huge confusions that are still to be resolved for understanding the taxonomy of the branched heterocytous forms as genus based clustering was almost not visible anywhere. This polyphyly of the branched cyanobacteria (within themselves) has already been indicated by previous works which had representative taxa of Indian origin (Gugger and Hoffmann 2004; Singh et al. 2013, 2014, 2015a, b). The results also reflected coherence with the findings of Komárek (2013) and Komárek et al. (2014) in anticipating more taxa from the branched heterocytous forms. It is also acknowledged again that the confusing morphology and close phenotypic characters certainly advocate the need of sustained polyphasic studies of the branched heterocytous forms (Jeeji-Bai 1972; Gugger and Hoffmann 2004; Kaštovský and Johansen 2008; Komárek and Mareš 2012; Dagan et al. 2013).
In the present work, we extended our molecular analyses by using the rbcL and nifD genes with an aim to create a robust database for the newly described species of Westiellopsis so that in the future, taxonomic studies focusing on this genus could get support from a well-standardized multilocus plan of work. In case of the rbcL gene, the closest phylogenetic affiliation was found with F. muscicola UTEX 1829 (AB075910) while the next closest relative was H. hibernicus BZ-3-1 (EU151924). In case of the nifD gene, the closest relative was Fischerella sp. UTEX 1903 (AY196955). It is notable that the nifD gene has also been used as a molecular marker for assessing cyanobacteria of both the unbranched heterocytous and the branched heterocytous forms (Singh et al. 2015b), while in few recent studies it has also been used as a marker for establishing the taxonomic identity of a new species Scytonema bilaspurensis Singh, sp. nov. (Singh et al. 2016). The use of the rbcL and the nifD genes proved to be futile in this case because of the dearth of proper sequences of the genus Westiellopsis and this indicates toward the database inconsistency.
The folding of the secondary structure of the 16S-23S ITS region again gave conclusive proof of the strain HPS being different from any other previously known heterocytous forms as comparisons with the secondary structure of two of the most closely related strains Westiellopsis sp. Ar73 (DQ786168) and Fischerella muscicola SAG 2027 (AM709634) revealed prominent differences. The analysis of the folding of the secondary structure of the ITS region also indicated that the strain HPS was indeed different from any known species of Westiellopsis known previously.
Thus, morphological, ecological, molecular, and phylogenetic interpretations when considered together gave enough taxonomic evidence that the strain HPS, isolated from a floating freshwater cyanobacterial mat is indeed a new addition to the genus Westiellopsis.
Taxonomic treatment
Westiellopsis ramosa Bagchi, sp. nov.—HOLOTYPE: India, Madhya Pradesh, Jabalpur, Ganga Sagar Lake, 23°10′N, 79°56′E (cryopreserved culture MCC 3176 deposited in Microbial Culture Collection (MCC), National Centre for Cell Science, Pune, India as Westiellopsis sp.; the culture was authenticated using 16S rRNA gene sequencing).
Etymology: Westiellopsis ramosa (ra.mo’sa. L. fem. adj. ramosa, having many branches, much-branched).
Description: Thallus greenish, free-floating, caespitose, composed of clusters of free, untangled filaments, not coalescent together. Main filaments not clearly torulous nor flexuous; no occurrence of biseriate filaments seen, prominently constricted at cross-walls; typical T-shaped branching. Branching usually unidirectional on one side of the filament. Cells in the main filaments vary in shapes; cylindrical to barrel shaped to being irregularly shaped; 5.21–9.21 μm in length and 4.03–6.50 μm in width. Branches usually thinner than the main filaments; constrictions more prominent at the basal parts from where the branches originate; later the cells are more cylindrical, non-constricted and slightly tapering at the apex. Cells in the branches usually rounded or irregularly rounded at the start while the later cells are more cylindrical with tapering ends; 6.85–9.78 μm in length and 2.86–3.65 μm in width. Heterocytes usually cylindrical in shape or sometimes appear slightly barrel shaped, may be both terminal and intercalary, and also show constrictions at the cross-walls 7.37–11.47 μm in length and 3.53–5.06 μm in width. Monocytes were observed rarely; are solitary and spherical; liberate singly from the branch endings; 4.64–5.42 μm in length and 4.98–5.29 μm in width; Akinetes not observed; reproduction usually by hormogonia, which may arise from the terminal cells or sometimes also from intercalary portions of secondary branches; monocytes may also sometimes escape from filaments and from chroococcoid stages which may serve as reproductive bodies.
Habitat: Freshwater.
Ecology of type locality: Average depth 58 m; eutrophic to dystrophic; temperature 21 °C; pH 7.1; phosphate 0.3 mg/L; NH4–N 4 mg/L; BOD 18 ppm; COD 28 ppm; dissolved oxygen 0.28 ppm; free carbon dioxide 28 ppm; total hardness 150 ppm; chloride 80 ppm, and total and calcium alkalinity 135, 70 ppm.
References
American Public Health Association (APHA) (2012) Standard methods for the examination of water and waste water. APHA, Washington, DC
Anagnostidis K, Komárek J (1990) Modern approach to classification system of cyanophytes 5-Stigonematales. Arch Hydrobiol Suppl Algol Stud 59:1–73
Bourrelly P (1970) Les algues d’eau douce. Initiation à la systématique, Tome III: Les Algues bleues et rouges, Les Eugléniens, Peridiniens et Cryptomonadines. Boubée & Cie, Paris
Bourrelly P (1984) Algues d’eau douce de la Nouvelle Calédonie recueillés par la Mission F. Starmuehlner en 1965 (Diatoméesexcluses).1 Partie: Cyanophycées, Rhodophycées, Xanthophycées, Pheophycées, Chlorophycées (sans Desmidiées). Rev Hydrobiol Trop 17:13–51
Castenholz RW (2001) Oxygenic photosynthetic bacteria. In: Boone DR, Castenholz RW (eds) Bergey’s manual of systematic bacteriology, vol 1. Springer, New York, pp 473–600
Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P, Gould SB, Goremykin VV, Rippka R, Marsac NTD, Gugger M, Lockhart PJ, Allen JF, Brune I, Maus I, Pühler A, Martin WF (2013) Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol 5:31–44. doi:10.1093/gbe/evs117
Desikachary TV (1959) Cyanophyta. ICAR monographs on algae. Indian Council of Agricultural Research, New Delhi
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucl Acids Res 17:7843–7853. doi:10.1093/nar/17.19.7843
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Molec Evol 17:368–376. doi:10.1007/BF01734359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Finsinger K, Scholz I, Serrano A, Morales S, Uribe-Lorio L, Mora M, Sittenfeld A, Weckesser J, Hess WR (2008) Characterization of true-branching cyanobacteria from geothermal sites and hotsprings of Costa Rica. Environm Microbiol 10:460–473. doi:10.1111/j.1462-2920.2007.01467.x
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416. doi:10.2307/2412116
Gkelis S, Rajaniemi P, Vardaka E, Moustaka-Gouni M, Lanaras T, Sivonen K (2005) Limnothrix redekei (Van Goor) Meffert (cyanobacteria) strains from lake Kastoria, Greece form a separate phylogenetic group. Microbial Ecol 49:176–182. doi:10.1007/s00248-003-2030-7
Gugger MF, Hoffmann L (2004) Polyphyly of true branching cyanobacteria (Stigonematales). Int J Syst Evol Microbiol 54:349–357. doi:10.1099/ijs.0.02744-0
Hoffmann L, Komárek J, Kaštovský J (2005) System of cyanoprokaryotes (cyanobacteria)–state. Algol Stud 117:95–115. doi:10.1127/1864-1318/2005/0117-0095
Janet M (1941) Westiellopsis prolifica, gen. et sp. nov., a New Member of the Stigonemataceae. Ann Bot (Oxford) 5:167–170
Jeeji-Bai N (1972) The genus Westiellopsis Janet. In: Desikachary TV (ed) Taxonomy and biology of blue-green algae. University of Madras, Madras, pp 62–74
Kaštovský J, Johansen JR (2008) Mastigocladus laminosus (Stigonematales, Cyanobacteria): phylogenetic relationship of strains from thermal springs to soil-inhabiting genera of the order and taxonomic implications for the genus. Phycologia 47:307–320. doi:10.2216/PH07-69.1
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi:10.1099/ijs.0.038075-0
Komárek J (2013) Cyanoprokaryota. 3. Heterocytous genera. In: Büdel B, Gärtner G, Krienitz L, Schagerl M (eds) Süswasserflora von Mitteleuropa/freshwater flora of Central Europe. Springer, Heidelberg
Komárek J (2016) Review of the cyanobacterial genera implying planktic species after recent taxonomic revisions according to polyphasic methods: state as of 2014. Hydrobiologia 764:259–270. doi:10.1007/s10750-015-2242-0
Komárek J, Mareš J (2012) An update to modern taxonomy (2011) of freshwater planktic heterocytous cyanobacteria. Hydrobiologia 698:327–351. doi:10.1007/s10750-012-1027-y
Komárek J, Kaštovský J, Mareš J, Johansen JR (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 86:295–335
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure culture of cyanobacteria. J Gen Microbiol 111:1–61. doi:10.1099/00221287-111-1-1
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol 4:406–425. doi:10.1093/oxfordjournals.molbev.a040454
Singh P, Singh SS, Mishra AK, Elster J (2013) Molecular phylogeny, population genetics and evolution of heterocystous cyanobacteria using nifH gene sequences. Protoplasma 250:751–764. doi:10.1007/s00709-012-0460-0
Singh P, Kaushik MK, Srivastava M, Mishra AK (2014) Phylogenetic analysis of heterocystous cyanobacteria (Subsections IV and V) using Highly Iterated Palindromes as molecular markers. Physiol Molec Biol Pl 20:331–342. doi:10.1007/s12298-014-0244-4
Singh P, Fatma A, Mishra AK (2015a) Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcL gene sequence data. Ann Microbiol 65:799–807. doi:10.1007/s13213-014-0920-1
Singh P, Singh SS, Aboal M, Mishra AK (2015b) Decoding cyanobacterial phylogeny and molecular evolution using an evonumeric approach. Protoplasma 252:519–535. doi:10.1007/s00709-014-0699-8
Singh P, Minj RA, Kunui K, Shaikh ZM, Suradkar A, Shouche YS, Mishra AK, Singh SS (2016) A new species of Scytonema isolated from Bilaspur, Chhattisgarh, India. J Syst Evol 54:519–527. doi:10.1111/jse.12202
Solarzano L (1969) Determination of ammonia in natural waters by the phenol hypochlorite method. Limnol Oceanogr 14:799–801
Strackerbrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Tomitani A (2004) Evolution of branching filamentous cyanobacteria: molecular-phylogenetic analyses of stigonematalean species. IFREE Report 2003–2004(2):1–5
Xia X (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Molec Biol Evol 30:1720–1728. doi:10.1093/molbev/mst064
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res 31:3406–3415. doi:10.1093/nar/gkg595
Acknowledgements
SNB & ND thank the Head Department of Biological Science Rani Durgavati University for extending the lab facilities. A part of the work was supported by the Department of Biotechnology (Department of Biotechnology; Grant no. BT/PR/0054/NDB/52/94/2007), Government of India, under the project ‘Establishment of Microbial Culture Collection’ and PS is thankful to the Department of Science and Technology (DST), India for the project YSS/2014/000879.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Karol Marhold.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Strains present in the compressed clusters in the 16S rRNA gene analysis of all the heterocystous cyanobacteria (PDF 442 kb)
Online Resource 2
Maximum likelihood (ML) tree of Westiellopsis ramosa sp. nov. based on the rbcL gene. Corresponding bootstrap values in the NJ and the MP trees are also indicated at the second and the third positions, respectively (PDF 124 kb)
Online Resource 3
Strains present in the compressed cluster in the rbcL analysis (PDF 418 kb)
Online Resource 4
Maximum likelihood (ML) tree of Westiellopsis ramosa sp. nov. based on the nifD gene. Corresponding bootstrap values in the NJ and the MP trees are also indicated at the second and the third positions, respectively (PDF 255 kb)
Online Resource 5
Strains present in the compressed cluster in the nifD analysis (PDF 104 kb)
Online Resource 6
16S rRNA alignment files with the strain HPS and all heterocystous cyanobacteria (NEX 98 kb)
Online Resource 7
16S rRNA alignment files with the strain HPS and only branched heterocystous cyanobacteria (NEX 233 kb)
Online Resource 8
rbcl alignment file with the strain HPS and all the considered taxa (NEX 49 kb)
Online Resource 9
nifD alignment file with the strain HPS and all the considered taxa (NEX 7 kb)
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Strains present in the compressed clusters in the 16S rRNA gene analysis of all the heterocytous cyanobacteria.
Online Resource 2. Maximum likelihood (ML) tree of Westiellopsis ramosa sp. nov. based on the rbcL gene.
Online Resource 3. Strains present in the compressed cluster in the rbcL analysis.
Online Resource 4. Maximum likelihood (ML) tree of Westiellopsis ramosa sp. nov. based on the nifD gene.
Online Resource 5. Strains present in the compressed cluster in the nifD analysis.
Online Resource 6. 16S rRNA alignment files with the strain HPS and all heterocytous cyanobacteria.
Online Resource 7. 16S rRNA alignment files with the strain HPS and only branched heterocytous cyanobacteria.
Online Resource 8. rbcL alignment file with the strain HPS and all the considered taxa.
Online Resource 9. nifD alignment file with the strain HPS and all the considered taxa.
Rights and permissions
About this article
Cite this article
Singh, P., Dubey, N. & Bagchi, S.N. Westiellopsis ramosa sp. nov., intensely branched species of Westiellopsis (cyanobacteria) from a freshwater habitat of Jabalpur, Madhya Pradesh, India. Plant Syst Evol 303, 1239–1249 (2017). https://doi.org/10.1007/s00606-017-1434-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1434-7