Abstract
Twenty-one populations (555 individuals) covering the entire native range of Pinus mugo Turra (dwarf mountain pine) were investigated for genetic variation scored at 13 nuclear microsatellite markers (nSSRs). The main objective of the present study was to determine the genetic structure across the present distribution of the species and locate populations of different genetic compositions. Most of the genetic variation was observed within the populations (95%). The assignment of populations based on Bayesian clustering methods revealed that the Sudeten populations of P. mugo form a separate genetic cluster. These stands have likely been established through the founder effects of Alpine migrants. The distribution and level of SSR polymorphisms, along with no evidence of isolation by distance or phylogeographic structure, indicate that the present populations of P. mugo have diverged relatively recently and originate from a larger glacial distribution of the species. One peripheral stand from Italy had the lowest values of most calculated genetic variation indices. This stand could therefore be more susceptible to genetic drift and a negative impact of predicted environmental changes. We discuss our findings with respect to previously published results on the genetic and morphological variation of P. mugo and with consideration for the conservation genetics of the species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polymorphisms observed in the genomes of various populations originate from demographic and evolutionary processes, such as gene flow, drift, mutations, recombination and selection (Nosil and Feder 2013). Across environmental landscapes, individuals experience various selective pressures that may result in differences observed among their genomes due to local adaptation (Schoville et al. 2012). Nonetheless, the genetic signatures of adaptive genetic variation (Lowry 2010) are different from neutral genetic variation resulting from population history, including long-term isolation and population range shifts.
The complexity of population history makes it challenging to elucidate the precise forces responsible for the genomic patterns of the polymorphisms observed in present populations. Studying the population genetics of forest tree species is a particularly formidable task, as most tree genomes are large. Forest tree species also have high genetic and phenotypic variation, large population sizes, low levels of linkage disequilibrium and long generation times. Moreover, forest tree research primarily focuses on a few species of high economic importance, and species that have an ecological value are generally ignored (Neale and Kremer 2011). Thus, for many species, the location of Pleistocene refugia and recolonization routes are difficult to trace back.
Undoubtedly, the population genetics of trees has recently gained widespread attention. Tree species have become useful models to study evolutionary processes in wild populations. Genomic approaches are constantly developed, and considerable effort has been placed into identifying genetic markers that will facilitate evolutionary studies and tree breeding. It is crucial to assess the genetic variation of present tree populations not only to determine which evolutionary factors have influenced populations during their evolution but also to preserve genetic resources, particularly in the face of ongoing environmental changes.
Pinus mugo Turra (dwarf mountain pine) is a European forest tree species that plays a key role in alpine areas, preventing avalanches and soil erosion. This species belongs to the P. mugo complex, a European group of closely related pine species that has a uniform genetic background but variable ecology, morphology and geographic distribution. In addition, the complex constitutes a good candidate for genetic studies of adaptive variation and speciation (Żukowska and Wachowiak 2016). P. mugo forms shrubs up to a few metres in height that inhabit the subalpine belt of mountain ranges in Europe (Critchfield and Little 1971). In the Alps, its range overlaps with another member of the P. mugo complex, Pinus uncinata (Ramond) Domin (mountain pine), a single-stemmed tree common in the Western Alps and the Pyrenees (Monteleone et al. 2006).
Studies on P. mugo have primarily concentrated on the morphology of its cones and needles (e.g. Boratyńska and Boratyński 2007; Boratyńska et al. 2004, 2014, 2015). Populations from the Eastern and Southern Carpathians differ morphologically from the stands in other locations. Furthermore, some anatomical traits of the needles exhibit adaptive and ecological variation. However, little is known about the genetic background underlying the phenotypic differentiation of P. mugo and the postglacial history of this species. Historical demographic processes in P. mugo cannot be inferred from palynological records, as the pollen of its trees greatly resembles the pollen of Pinus sylvestris L. and Pinus nigra Arn. (Willis et al. 1998). Macrofossil data are also insufficient to distinguish these species (García-Amorena et al. 2007). Mitochondrial DNA, which is maternally inherited in conifers, has a relatively high mutation rate, which is sufficient to study recent divergence events (Sinclair et al. 1998). Unfortunately, variations in mtDNA genetic markers originally developed for P. sylvestris (Soranzo et al. 2000) are also too weak to make significant inferences regarding postglacial history of P. mugo (Wachowiak et al. 2013). A few studies using chloroplast microsatellite markers and/or isozymes have demonstrated the high genetic diversity accompanied by low interpopulation differentiation of P. mugo (Lewandowski et al. 2000; Slavov and Zhelev 2004; Heuertz et al. 2010; Sannikov et al. 2011; Dzialuk et al. 2012; Boratyńska et al. 2014). Higher differentiation among mountain ranges than within these areas was also shown (Dzialuk et al. 2012; Boratyńska et al. 2014). However, these studies were restricted to narrow areas of the present range of P. mugo.
The aim of the present study was to assess the level and distribution of neutral genetic variation across the native range of P. mugo. We used a set of 13 nuclear simple sequence repeat (nSSR) markers to examine genetic relationships within and among the studied populations and evaluate their phylogeographic structure. We also assessed whether populations occupying different mountain ranges have similar or different genetic compositions, i.e. whether they share a recent history or whether they are genetically isolated. To our knowledge, the genetic variation of P. mugo has not yet been addressed using nSSR markers. Furthermore, the present study encompasses all parts of the present geographic range of P. mugo. The results are discussed in terms of previous genetic research and their influence on the conservation genetics of P. mugo.
Materials and methods
Plant material and DNA extraction
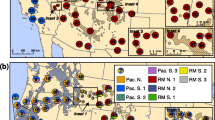
We analysed 21 populations of P. mugo, with 24–30 individuals per population, yielding a total of 555 trees studied (Fig. 1; Table 1). The populations cover the native range and putative glacial refugia of the species. The collected samples were dried at 37 °C and ground in a MM 400 Mixer Mill (Retsch, Germany) prior to DNA extraction. Genomic DNA was extracted from 50–100 mg of needle tissue, according to Dumolin et al. (1995). The final incubation was conducted using RNase A at 60 °C for 30 min. The DNA concentration was measured using a BioPhotometer (Eppendorf AG, Germany) and adjusted to 15 ng/μl.

(modified after Critchfield and Little (1971))
Map showing the genetic structure and approximate geographic location of the Pinus mugo populations analysed in the present study. Bayesian clustering revealed two genetic clusters (I: SUD1–4; and II: A1–7, CAR1–4, APN, BAL1–5). Pie charts show the probability of belonging to one of the two genetic clusters. The range of the species is shaded in grey
Molecular analysis
We selected 13 nSSRs that provided repeatable, unambiguous bands of expected sizes with sufficient polymorphisms from the set of 22 nSSRs initially screened on our plant material (Online Resource 1). The selected loci were amplified in three multiplex PCRs, each in a total volume of 10 μl, using the Qiagen Multiplex PCR Kit (Qiagen, Germany) in Applied Biosystems Veriti and 2720 thermal cyclers (Life Technologies, USA). The PCRs contained 3 μl of template DNA, 1× Qiagen Multiplex PCR Master Mix, 0.5× Q-Solution and 0.05–0.1 μM each of forward and reverse primer. A specific fluorescent dye was attached to the forward primer in each primer pair. The standard amplification procedure for each of the three multiplexes started with initial denaturation at 95 °C for 15 min and varied as follows: (1) psyl2, psyl16, psyl18, psyl25, psyl36, psyl42, psyl44 and psyl57 were amplified in Multiplex I with 38 cycles (30-s denaturation at 94 °C, 90-s annealing at 55 °C and 90-s extension at 72 °C); (2) ptTX2146 and SPAG 7.14 were amplified in Multiplex II with 35 cycles (30-s denaturation at 94 °C, 90-s annealing at 56 °C and 90-s extension at 72 °C); and (3) ptTX3025, ptTX4001 and ptTX4011 were amplified in Multiplex III with 10 cycles of touchdown (30-s denaturation at 94 °C, 40-s annealing at 65 °C Δ↓ 1 °C and 60-s extension at 72 °C), followed by 32 cycles (30-s denaturation at 94 °C, 60-s annealing at 55 °C and 60-s extension at 72 °C). The final extension at 72 °C lasted 15 min for Multiplex I and II and 7 min for Multiplex III. The PCR products and the GeneScan 500 LIZ Size Standard (Life Technologies, USA) were separated on a capillary sequencer using the Applied Biosystems 3130 Genetic Analyser (Life Technologies, USA). The alleles were determined based on their sizes using the GeneMapper Software ver. 4.0 (Life Technologies, USA). The raw data were manually assessed and converted into discrete allele sizes using the automated binning software Tandem (Matschiner and Salzburger 2009).
Tests for genetic equilibrium at the loci
The score test (U test for heterozygote deficit, Raymond and Rousset 1995), implemented in GENEPOP ver. 4.3 (Rousset 2008), was used to assess deviations from Hardy–Weinberg equilibrium (HWE) for each P. mugo population. Additionally, the multiple sample score test was used for each locus across all populations (Raymond and Rousset 1995). Whenever significant deficiencies of heterozygotes were observed, we suspected the presence of relatively high frequency of null alleles. As null alleles may significantly overestimate population differentiation as a result of the presence of false homozygotes (Chapuis and Estoup 2007), we assessed whether our data set contained null alleles using Micro-Checker software ver. 2.2.3 (Van Oosterhout et al. 2004). In a subsequent analysis, we calculated the inbreeding coefficients for each population using INEst software ver. 2.0 (Chybicki 2015). The calculations were performed using a Bayesian approach, 500,000 Markov chain Monte Carlo (MCMC) iterations with the thinning parameter set at 500 and burning set to 50,000 cycles. We selected the full model, considering the presence of both null alleles and inbreeding and potential genotyping failures (F ISnull). To verify the significance of inbreeding, we repeated the analysis using a random mating model (i.e. assuming that F = 0; F IS) and performed the Bayesian procedure of model comparison. Finally, we determined which model (full model = F ISnull vs. random mating model = F IS) fitted better to the data by computing deviance information criterion (DIC) for each option. In addition, the exact test for genotypic disequilibrium between all pairs of loci in each population was performed using GENEPOP ver. 4.3 with the following settings: MCMC dememorization number set at 10,000, 100 batches and 5000 iterations per batch.
Genetic diversity and differentiation
Within-population genetic diversity was assessed with the following parameters: the mean number of alleles (A N), mean effective number of alleles (A E), number of private alleles (i.e. unique to a single population; A P), and observed and unbiased expected heterozygosity (H O and uHE, respectively), using GenAlEx software ver. 6.5 (Peakall and Smouse 2006). The mean rarefied allelic richness across all loci (A R24), based on a minimum sample size of 24 diploid individuals, was calculated with FSTAT ver. 2.9.3.2 (Goudet 2001). The statistical significance of the differences in the values of the calculated genetic indices between the Sudeten versus the other stands (groups defined by the Bayesian clustering; see ‘Results’ section) was calculated using Student’s t test implemented in JMP Pro ver. 12.1.0 software.
To estimate the proportion of the overall genetic variation resulting from differentiation among the P. mugo populations, we computed unbiased global and pairwise F ST using FreeNA software (Chapuis and Estoup 2007). FreeNA considers the potential bias reflecting the presence of null alleles using the Excluding Null Alleles (ENA) correction method (F STnull). A total of 10,000 replicates were set to calculate the bootstrap 95% confidence interval (95% CI) for the global and pairwise F ST and F STnull.
Population structure
The genetic structure of the P. mugo populations was determined using the Bayesian clustering approach implemented in STRUCTURE ver. 2.3 (Pritchard et al. 2000). This software uses multilocus genotype data to assign individuals to genetically divergent clusters. The settings in STRUCTURE were as follows: a burn-in period of 50,000 followed by 500,000 iterations using the correlated allele frequency model and admixture ancestry model. A total of 20 independent runs were set for each potential number of clusters (K = 1–21). The output from STRUCTURE was visualized in STRUCTURE HARVESTER (Earl and von Holdt 2011), and the optimum value of K was determined according to the Evanno method (Evanno et al. 2005). Multiple runs for the optimum K value were aligned using CLUMPP ver. 1.1.2 (Jakobsson and Rosenberg 2007). Additionally, we conducted the hierarchical analysis of molecular variance (AMOVA) to assess the level of differentiation among the following groups: (a) all populations; (b) groups of populations divided into mountain regions according to their geographic locations; and (c) groups defined by the Bayesian clustering. The analysis was performed using both F ST and R ST distance type in GenAlEx ver. 6.5.
Isolation by distance and phylogeographic structure
A Mantel test was performed to determine whether differentiation among the P. mugo populations results from isolation by distance (IBD). Specifically, we tested the correlation between the matrices of pairwise geographic distances (logarithmic scale) and pairwise population F STnull with 1000 permutations as implemented in GenAlEx ver. 6.5.
The potential existence of phylogeographic structure was assessed with the permutation test according to Hardy et al. (2003). To that end, we computed and compared pairwise R ST and permuted R ST (pR ST) with 10,000 permutations using the SPAGeDI software ver. 1.5 (Hardy and Vekemans 2002). R-statistics is an analogue to F-statistics, but it is allele-size-based, as it considers not only the identity of the alleles (as F-statistics) but also the differences among their sizes. Thus, R ST reflects interpopulation differences resulting from genetic drift and mutation processes according to the stepwise mutation model (SMM). When R ST is significantly higher than pR ST, allele sizes contribute to the observed genetic differentiation, indicating the existence of phylogeographic structure (Hardy et al. 2003).
Considering potential population size fluctuations, we assessed whether the P. mugo stands analysed in the present study experienced a genetic bottleneck. Such populations tend to lose rare alleles and experience a reduction in heterozygosity. However, the allelic diversity decreases faster than heterozygosity; thus, the observed heterozygosity is greater than that expected from the observed number of alleles (Luikart and Cornuet 1998). We selected a one-tailed Wilcoxon’s sign-rank test to examine departures from HWE using the Bottleneck ver. 1.2.02 software (Cornuet and Luikart 1996). This test is considered the most powerful approach to examine heterozygosity excess (Peery et al. 2012). The analysis was conducted under both the pure SMM model and the two-phase model (TPM), enabling single-step and multi-step mutations. TPM is considered the most suitable for most microsatellite markers, as these markers do not mutate under strict SMM (Di Rienzo et al. 1994). The TPM option was set to incorporate 70% of the single-step mutations and 30% of the multi-step changes.
Results
Polymorphism at microsatellite loci
All 13 microsatellite loci included in the final data set were polymorphic. The degree of genetic diversity calculated in the present study was highly dependent on the set of loci, as the ‘psyl’ markers were less polymorphic than the ‘ptTX’ and ‘SPAG’ loci. In total, we detected 133 alleles, yielding a mean number of ~10 alleles per locus. We detected 15 alleles that are specific to particular P. mugo stands, with a frequency varying from 0.017 to 0.067. These alleles were identified in populations from different mountain regions for seven nSSRs (Online Resource 2).
The mean number of alleles per population (AN) was equal to 4.98 and did not significantly vary across all populations of P. mugo, except for A5 (A N = 3.76). In addition, the frequency of many alleles was low; thus, the number of alleles that had a major influence on the level of genetic variation (effective number of alleles, A E) was lower than A N (mean A E = 2.64). The values of allelic richness (A R24) were only slightly lower than A N (mean A R24 = 4.89). Therefore, the variation in the number of individuals sampled from a particular population did not affect the degree of A N (Table 2).
The results obtained using Micro-Checker indicated that null alleles were present in 10 loci with frequencies between ~0.03 for psyl42 to ~0.14 for ptTX4011. However, the levels of observed heterozygosity (H O) for particular populations (range 0.33–0.52, average 0.40) were only slightly lower than the values of unbiased expected heterozygosity (uH E, equivalent to gene diversity) (range 0.35–0.53, average 0.44). The values of inbreeding coefficient with null alleles correction (F ISnull) were generally low and significantly different from zero only in some populations (Table 2). We concluded that inbreeding did not have a relevant influence on genetic diversity, and deviations from HWE at some loci primarily reflected the presence of null alleles. The frequency of null alleles in our data set was lower than the threshold value of 0.19 (Chapuis et al. 2008), above which the value of HE is significantly underestimated due to null alleles. Therefore, all 13 loci were used in further analyses.
Evidence of linkage disequilibrium between some pairs of loci was detected in populations A5 (psyl57 vs. ptTX2146), A7 (psyl2 vs. psyl25, psyl2 vs. psyl36 and psyl25 vs. psyl36) and BAL2 (psyl16 vs. psyl57, psyl57 vs. SPAG 7.14 and ptTX4001 vs. ptTX4011) (p < 0.001).
Genetic variation within populations
On average, we observed 4.98 different alleles per locus for each population. The mean value of A R24 was 4.89. The population from Monte Baldo in Italy (A5) exhibited the lowest values of genetic diversity of all tested P. mugo stands for most parameters (A N = 3.76, A E = 2.10, A R24 = 3.74, no private alleles). HO varied from 0.33 for SUD2 to 0.52 for A2 (mean H O = 0.40). This value was slightly lower than uHE, with values between 0.35 for SUD2 and 0.53 for A7 (mean uHE = 0.44) (Table 2). The values of H O and H E differed across loci from 0.07 and 0.10 for psyl18 (for H O and H E, respectively) to 0.79 and 0.89 for SPAG 7.14 (data not shown). In addition, the differences between the Sudeten and the remaining populations were statistically significant for A N, A R24, H O and uHE (Student’s t test; p < 0.05). We observed a weak inbreeding across the P. mugo populations (F ISnull ranging from 0.01 to 0.12, average 0.06), which was significantly different from zero only in a subset of the populations, namely in SUD3, A1, A5, CAR1, CAR2, APN and BAL5 (Table 2). In other samples, 95% CI overlapped zero.
Differentiation among populations
Most of the genetic diversity was observed within the studied populations. The global level of genetic differentiation among the P. mugo stands calculated using FreeNA software (F ST) was low and equal to 0.051 (95% CI 0.034–0.069). The value of F STnull, i.e. using the Excluding Null Alleles correction method, was 0.052 (95% CI 0.037–0.070). Almost all pairwise population values of F STnull were significantly greater than zero and varied from 0.008 between A3 and CAR2 to 0.156 between SUD2 and A7 (Online Resource 3). The F ST/F STnull values per locus ranged between 0.012/0.023 for ptTX3025 and 0.116/0.114 for ptTX4011 (data not shown).
Population structure
Based on the results of the Bayesian clustering, the P. mugo populations were clearly separated into two clusters using STRUCTURE according to the method described by Evanno et al. (2005). The four Sudeten populations (SUD1-4) were assigned to the first genetic cluster, whereas all the remaining stands from the other mountain ranges formed the second cluster (Figs. 1, 2). The AMOVA results using the F ST distance indicated that 5% of the variance was among the studied P. mugo populations. The remaining 95% reflected the differentiation observed within populations. When divided into mountain regions (see Table 1), the analysis showed that only 1% of the genetic variance reflected differences among the mountain regions, whereas 4% of the genetic variance among the P. mugo populations reflected differences within the regions. Furthermore, when we separated the populations into two groups detected in the Bayesian clustering (SUD vs. the other stands), the variance was 3% between the groups and 4% among the populations within the groups (all results significant at p < 0.001) (Table 3). When we performed the analysis on the R ST distance, only 3% of variance reflected the differentiation among populations (p < 0.001). These results were not significant when the analysis was performed on the mountain regions or groups obtained using Bayesian clustering.
Isolation by distance and phylogeographic structure
The Mantel test yielded no significant correlation (R = 0.070; p = 0.153). Similarly, the results of the permutation test did not support the existence of phylogeographic structure among the P. mugo populations analysed in the present study (R ST = 0.037 < pR ST = 0.043; p = 0.649). Therefore, neither the SMM nor IBD could explain the genetic differentiation observed in our data set. Finally, no signs of bottlenecks were observed either under the SMM or TPM model in any population.
Discussion
Distinct genetic composition of Pinus mugo in the Sudetes
The present study revealed that P. mugo in the Sudetes differs from other populations of this species. Notably, in the past, large areas of P. mugo in the Sudetes were replaced with pastures from the fifteenth century until the end of the nineteenth century. P. mugo naturally recovered, at least partially, particularly on the abrupt slopes of the glacial cirques, but this species may have been planted in certain places, such as the Czech Republic (Sobierajska et al. 2010). Therefore, the origin of some of the present Sudeten populations of P. mugo is uncertain. Nevertheless, morphological studies showed that populations on the plateaux do not differ from those inhabiting the glacial cirques (Sobierajska and Boratyńska 2008; Sobierajska et al. 2010). Thus, even if some parts of the Polish Sudetes were artificially reforested, this restoration was conducted most likely with the seeds of local origin (Sobierajska and Boratyńska 2008; Sobierajska et al. 2010).
Dzialuk et al. (2012) suggested that P. mugo survived the Last Glacial Maximum in different refugia, namely in the Alps, Sudetes and Carpathians. In morphological surveys, however, the Sudeten populations are only slightly different compared with the other mountain regions (Boratyńska and Boratyński 2007; Boratyńska et al. 2015). We hypothesize that the Polish Sudeten populations of P. mugo are younger than the populations occupying other mountain ranges. According to the ‘leading edge’ model of colonization, these species may have been established via the founder effects (Hampe and Petit 2005) of Alpine migrants. The slightly lower genetic diversity (i.e. the values of A N, A R24, H O and uHE) of the Sudeten populations compared with the average supports this idea. Orsini et al. (2013) argued that patterns of genetic variation should be interpreted with caution, as neutral genetic variation may be determined not only via dispersal limitation but also through colonization history and local adaptation. In the present study, we assume isolation based on a colonization (IBC) scenario, under which populations are expected to differentiate at neutral loci, but this differentiation does not correlate with geographic or ecological distance (Orsini et al. 2013). The differentiation between the Sudeten populations and other stands and lack of IBD signal is consistent with this scenario. The standing genetic variation present in the founding population of P. mugo was likely sufficient to facilitate rapid genetic adaptation to the local environment in the Sudetes. As a consequence, the newly established populations may have rapidly grown in size. Even if we assume a free gene exchange between the Sudetes and Alps, a simple numerical advantage of the first migrants led to the pattern of genetic differentiation. The rate of gene flow between the Sudetes and Alps has been too slow, or simply, there has not been enough time to eliminate this long-lasting founder effect.
Differentiation among populations and mountain regions
We observed that populations of P. mugo are differentiated only to a small extent (F STnull = 0.052) and that variation is primarily distributed within the populations. The classical value of F ST (without the correction of null alleles) was equal to 0.051. Therefore, the occurrence of null alleles did not affect the level of F ST. Indeed, high within-population diversity accompanied with little interpopulation differentiation at neutral markers is expected for highly outcrossing species such as conifers (Hamrick and Godt 1996).
The Mantel test performed on our data set was insignificant. Thus, IBD was not a mechanism shaping the genetic structure of P. mugo populations. The global genetic differentiation that considered allele sizes (R ST) was lower than the F ST value. The results of the permutation test were also insignificant (R ST = 0.037 < pR ST = 0.043; p = 0.649). This finding indicates the absence of phylogeographic structure and that SMM did not play a significant role in shaping the genetic structure observed in the data set obtained in the present study.
Previous work on P. mugo yielded similar or higher values of interpopulation genetic differentiation (F ST = 0.041 for isoenzymes, Slavov and Zhelev 2004; F ST = 0.076 and G ST = 0.070 for cpSSRs, Heuertz et al. 2010; F ST = 0.069 in the Carpathians and F ST = 0.033 in the Alps for isoenzymes, Sannikov et al. 2011; F ST = 0.082 and R ST = 0.137 for cpSSRs, Dzialuk et al. 2012; G ST = 0.020 for isoenzymes and G ST = 0.017 for cpSSRs, Boratyńska et al. 2014). Moreover, Dzialuk et al. (2012) observed a high level of differentiation among the mountain ranges (the Sudetes, Alps and Carpathians) using cpSSRs (11.86% for F ST and 19.64% for R ST). These authors suggested that this finding reflected ancient fragmentation and the long isolation of P. mugo stands. The genetic differentiation among the mountain regions calculated in the present study was considerably lower (1% for F ST and insignificant for R ST), as the variation primarily reflected differentiation within the populations (95% for F ST and insignificant for R ST) and among the populations within the regions (4% for F ST and insignificant for R ST). Higher degrees of genetic differentiation may reflect the fact that the mentioned study was based on different markers (cpSSRs) and conducted on a much smaller area of the P. mugo range compared with the present study.
Morphological studies of P. mugo showed that populations from the Eastern and Southern Carpathians differ from the other stands of P. mugo (Boratyńska et al. 2004, 2015). This result indicates that these populations may have originated from a different Pleistocene refugium or reflect the adaptive significance of needle features. The results of the present study rule out the first option. The weak genetic differentiation, lack of IBD signal and the lack of phylogeographic structure in the present study suggest the relatively recent fragmentation of a historically larger glacial P. mugo range. As the species alternately spread and retreated during cold and warm periods of the Pleistocene (Willis et al. 2000; Latałowa et al. 2004), populations from distinct mountain ranges may have come into contact, thereby mixing the initial gene pools. The isolation of particular stands in the most elevated mountains likely started only 8000–9000 years ago (Boratyńska et al. 2004). Hence, there is a weak genetic differentiation of P. mugo. Previous genetic studies indeed identified a distinct origin of populations of conifers inhabiting major mountain ranges (Vendramin et al. 1999; Afzal-Rafii and Dodd 2007), but other studies did not reveal the strong genetic divergence of mountain isolates (Robledo-Arnuncio et al. 2005), similar to the results of the present study.
Implications for conservation genetics of Pinus mugo
In our study, most parts of the analysed range of P. mugo (the Alps, Carpathians, Apennines and Balkans) showed no evidence of genetic differentiation. Consequently, based on the results of the nSSRs analysis, these populations should be treated as a single conservation unit. However, populations from the Sudetes, which are located in the northern limits of the natural range of P. mugo, showed distinct genetic structure, so they should be considered as a separate genetic group. Additionally, in the present study, one of the Italian populations (A5) had the lowest values for most calculated genetic variation indices, even lower than the most isolated stand from the Apennines (APN). This population inhabits the south-western-most area of the P. mugo range. We even observed that four genotypes were repeated twice in this stand. Nevertheless, no evidence of a recent genetic bottleneck was observed in this population or in any other studied location. Nevertheless, considering its evidently lower genetic diversity, A5 may be more prone to the negative effect of genetic drift. Maintaining high within-population genetic diversity is particularly important to adapt to a changing environment (Markert et al. 2010). P. mugo is an important component of alpine landscapes, as its trees prevent avalanches and soil erosion. Climate warming is likely to cause migration of P. mugo to higher altitudes, similarly to the retreats observed during warmer interglacials (Heuertz et al. 2010). As the response of various species to environmental changes will most likely depend on the reaction of populations located at range margins (Hampe and Petit 2005), it is important to obtain comprehensive genetic information, particularly for peripheral populations of P. mugo, to properly formulate management strategies for the conservation of genetic resources of this species.
Conclusions
It appears that present populations of P. mugo originate from a larger distribution of the species. They have most likely been isolated for a relatively short period of time, not long enough to cause pronounced genetic differentiation among them. Therefore, the location of the ancient refugial populations of P. mugo remains poorly recognized. The data obtained in the present study confirm the potential northward movement of P. mugo during the postglacial recolonization of Europe. Specifically, the Sudeten populations may have been established through the founder effects of migrants from the Alps. We also identified a single peripheral population of P. mugo that should be given particular attention regarding the preservation of the genetic resources of the species.
References
Afzal-Rafii Z, Dodd RS (2007) Chloroplast DNA supports a hypothesis of glacial refugia over postglacial recolonization in disjunct populations of black pine (Pinus nigra) in Western Europe. Molec Ecol 16:723–736. doi:10.1111/j.1365-294X.2006.03183.x
Boratyńska K, Boratyński A (2007) Taxonomic differences among closely related pines Pinus sylvestris, P. mugo, P. uncinata, P. rotundata and P. uliginosa as revealed in needle sclerenchyma cells. Flora 202:555–569. doi:10.1016/j.flora.2006.11.004
Boratyńska K, Muchewicz E, Drojma M (2004) Pinus mugo Turra geographic differentiation based on needle characters. Dendrobiology 51:9–17
Boratyńska K, Dzialuk A, Lewandowski A, Marcysiak K, Jasińska AK, Sobierajska K, Tomaszewski D, Burczyk J, Boratyński A (2014) Geographic distribution of quantitative traits variation and genetic variability in natural populations of Pinus mugo in Central Europe. Dendrobiology 72:65–84. doi:10.12657/denbio.072.006
Boratyńska K, Jasińska AK, Boratyński A (2015) Taxonomic and geographic differentiation of Pinus mugo complex on the needle characteristics. Syst Biodivers 13:581–595. doi:10.1080/14772000.2015.1058300
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Molec Biol Evol 24:621–631. doi:10.1093/molbev/msl191
Chapuis MP, Lecoq M, Michalakis Y, Loiseau AG, Sword G, Piry S (2008) Do outbreaks affect genetic population structure? A worldwide survey in Locusta migratoria, a pest plagued by microsatellite null alleles. Molec Ecol 17:3640–3653. doi:10.1111/j.1365-294X.2008.03869.x
Chybicki I (2015) INEST 2.0. Available at: http://www.ukw.edu.pl/pracownicy/strona/igor_chybicki/software_ukw/
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Critchfield WB, Little EL (1971) Geographic distribution of the pines of the world. U.S. Department of Agriculture, Washington
Di Rienzo AA, Peterson C, Garza JC, Valdes AM, Slatkin M, Freime RB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256. doi:10.1007/BF00220937
Dzialuk A, Boratyński A, Boratyńska K, Burczyk J (2012) Geographic patterns of genetic diversity of Pinus mugo (Pinaceae) in Central European mountains. Dendrobiology 68:31–41
Earl DA, von Holdt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resources 4:359–361. doi:10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molec Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
García-Amorena I, Gómez Manzaneque F, Rubiales JM, Granja HM, Soares de Carvalho G, Morla C (2007) The Late Quaternary coastal forests of western Iberia: a study of their macroremains. Palaeogeogr Palaeoclimatol Palaeoecol 254:448–461. doi:10.1016/j.palaeo.2007.07.003
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at: http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from Goudet (1995)
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467. doi:10.1111/j.1461-0248.2005.00739.x
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans Ser B Biol Sci 351:1291–1298
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molec Ecol Notes 2:618–620. doi:10.1046/j.1471-8286.2002.00305.x
Hardy OJ, Charbonnel N, Fréville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Heuertz M, Teufel J, González-Martínez SC, Soto A, Fady B, Alía R, Vendramin GG (2010) Geography determines genetic relationships between species of mountain pine (Pinus mugo complex) in western Europe. J Biogeogr 37:541–556. doi:10.1111/j.1365-2699.2009.02223.x
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Latałowa M, Tobolski K, Nalepka D (2004) Pinus L. subgenus Pinus (subgen. Diploxylon (Koehne) Pilger)—Pine. In: Ralska-Jasiewiczowa M (ed) Late glacial and Holocene history of vegetation in Poland based on isopollen maps. W. Szafer Institute of Botany, Kraków, pp 165–177
Lewandowski A, Boratyński A, Mejnartowicz L (2000) Allozyme investigations on the genetic differentiation between closely related pines—Pinus sylvestris, P. mugo, P. uncinata, and P. uliginosa (Pinaceae). Pl Syst Evol 221:15–24. doi:10.1007/BF01086377
Lowry DB (2010) Landscape evolutionary genomics. Biol Lett 6:502–504. doi:10.1098/rsbl.2009.0969
Luikart G, Cornuet J-M (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biol 12:228–237
Markert JA, Champlin DM, Gutjahr-Gobell R, Grear JS, Kuhn A, McGreevy TJ Jr, Roth A, Bagley MJ, Nacci DE (2010) Population genetic diversity and fitness in multiple environments. BMC Evol Biol 10:205. doi:10.1186/1471-2148-10-205
Matschiner M, Salzburger W (2009) TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics 25:1982–1983. doi:10.1093/bioinformatics/btp303
Monteleone I, Ferrazzini D, Belletti P (2006) Effectiveness of neutral RAPD markers to detect genetic divergence between the subspecies uncinata and mugo of Pinus mugo Turra. Silva Fenn 40:391–406
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122. doi:10.1038/nrg2931
Nosil P, Feder JL (2013) Genome evolution and speciation: toward quantitative descriptions of pattern and process. Evolution 67:2461–2467. doi:10.1111/evo.12191
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Molec Ecol 22:5983–5999. doi:10.1111/mec.12561
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molec Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Robinson S, Vásquez-Carrillo C, Pauli JN, Palsbøll PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Molec Ecol 21:3403–3418. doi:10.1111/j.1365-294X.2012.05635.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Robledo-Arnuncio JJ, Collada C, Alía R, Gil L (2005) Genetic structure of montane isolates of Pinus sylvestris L. in a Mediterranean refugial area. J Biogeogr 32:595–605. doi:10.1111/j.1365-2699.2004.01196.x
Rousset F (2008) Genepop’007: a complete re-implementation of the Genepop software for Windows and Linux. Molec Ecol Resources 8:103–106. doi:10.1111/j.1471-8286.2007.01931.x
Sannikov SN, Petrova IV, Schweingruber F, Egorov EV, Parpan TV (2011) Genetic differentiation of Pinus mugo Turra and P. sylvestris L. populations in the Ukrainian Carpathians and the Swiss Alps. Russ J Ecol 42:270–276. doi:10.1134/s1067413611040151
Schoville SD, Bonin A, François O, Lobreaux S, Melodelima C, Manel S (2012) Adaptive genetic variation on the landscape: methods and cases. Annual Rev Ecol Evol Syst 43:23–43. doi:10.1146/annurev-ecolsys-110411-160248
Sinclair WT, Morman JD, Ennos RA (1998) Multiple origins for Scots pine (Pinus sylvestris L.) in Scotland evidence from mitochondrial DNA variation. Heredity 80:233–240. doi:10.1046/j.1365-2540.1998.00287.x
Slavov GT, Zhelev P (2004) Allozyme variation, differentiation, and inbreeding in populations of Pinus mugo in Bulgaria. Canad J Forest Res 34:2611–2617. doi:10.1139/x04-127
Sobierajska K, Boratyńska K (2008) Variability of needle characteristics of Pinus mugo Turra populations in the Karkonosze Mountains in Poland. Dendrobiology 59:41–49
Sobierajska K, Boratyńska K, Marcysiak K (2010) Variation of cone characters in Pinus mugo (Pinaceae) populations in the Giant Mountains (Karkonosze, Sudetes). Dendrobiology 63:33–41
Soranzo N, Alía R, Provan J, Powell W (2000) Patterns of variation at a mitochondrial sequence-tagged-site locus provides new insights into the postglacial history of European Pinus sylvestris populations. Molec Ecol 9:1205–1211. doi:10.1046/j.1365-294x.2000.00994.x
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Molec Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Vendramin GG, Degen B, Petit RJ, Anzidei M, Madaghiele A, Ziegenhagen B (1999) High level of variation at Abies alba chloroplast microsatellite loci in Europe. Molec Ecol 8:1117–1126. doi:10.1046/j.1365-294x.1999.00666.x
Wachowiak W, Boratyńska K, Cavers S (2013) Geographical patterns of nucleotide diversity and population differentiation in three closely related European pine species in the Pinus mugo complex. Bot J Linn Soc 172:225–238. doi:10.1111/boj.12049
Willis KJ, Bennett KD, Birks HJB (1998) The late Quaternary dynamics of pines in Europe. In: Richardson DM (ed) Ecology and biogeography of Pinus. Cambridge University Press, Cambridge, pp 107–121
Willis KJ, Rudner E, Sümegi P (2000) The full-glacial forests of Central and Southeastern Europe. Quatern Res 53:203–213. doi:10.1006/qres.1999.2119
Żukowska WB, Wachowiak W (2016) Utility of closely related taxa for genetic studies of adaptive variation and speciation: current state and perspectives in plants with focus on forest tree species. J Syst Evol 54:17–28
Acknowledgements
The authors would like to thank K. Boratyńska and A. Boratyński for the collection of the plant material and M. Litkowiec for technical support and valuable comments concerning the data analysis. The work was financially supported through a grant from the Polish National Science Centre (Grant No. DEC-2012/05/E/NZ9/03476).
Funding
This study was funded through a grant from the Polish National Science Centre (Grant No. DEC-2012/05/E/NZ9/03476).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Handling editor: Andreas Tribsch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. Characteristics of the 22 nSSR markers initially tested.
Online Resource 2. Private alleles detected in the analysed Pinus mugo populations.
Online Resource 3. Pairwise matrix of F ST Excluding Null Alleles (F STnull) among the Pinus mugo populations.
Rights and permissions
About this article
Cite this article
Żukowska, W.B., Wachowiak, W. Nuclear microsatellite markers reveal the low genetic structure of Pinus mugo Turra (dwarf mountain pine) populations in Europe. Plant Syst Evol 303, 641–651 (2017). https://doi.org/10.1007/s00606-017-1395-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1395-x