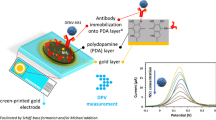
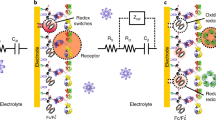
Abstract
Non-structural 1 (NS1) is a protein biomarker that can be found in blood in the early stages of dengue and related infections (Zika and Chikungunya). This study aims to develop a biosensor to selectively quantify NS1 using DNA aptamer co-immobilized on gold electrodes with 6-(ferrocenyl)hexanethiol (FCH) using electrochemical capacitive spectroscopy. This technique uses a redox probe (FCH) immobilized on the self-assembled monolayer to convert impedance into capacitance information. The developed platform was blocked with bovine serum albumin before NS1 exposure and the ratio between aptamers and FCH was optimized. The aptasensor was tested using commercial NS1 serotype 4 in phosphate-buffered saline and commercial undiluted human serum. Using the optimum applied potential provides high sensitivity (3 and 4 nF per decade) and low limit of detection (30.9 and 41.8 fg/mL) with a large linear range (10 pg to 1 µg/mL and 10 pg to 100 ng/mL, respectively). Both results exhibit a residual standard deviation value < 1%. The results suggested that this aptasensor was capable of detecting NS1 in the clinical range and can be applied to any other specific aptamer with FCH, opening the path for label-free miniaturized point-of-care devices with high sensitivity and specificity.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the World Health Organization (WHO), dengue has been identified as one of the ten threats to global health in 2019 [1]. This disease has become a main public health concern over last years, overcoming the status of neglected tropical disease [2, 3]. Dengue is transmitted by Aedes mosquitoes [4], infected by Flaviviridae virus (DENV) [5], and has four different serotypes (DENV-1, DENV-2, DENV-3, and DENV-4), which makes it difficult to create a tetravalent vaccine [2]. Over 3 million cases occurred in Brazil, only in 2022 [6]. To change this situation, early diagnosis is required with low cost, fast procedures, and high selectivity.
Dengue virus leads the infected cells to produce ten proteins: three structural proteins (capsid, membrane, and enveloped glycoprotein, or protein E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) [4]. Between them, NS1 is the only glycoprotein, with approximately 46–50 kDa, secreted from all serotypes [7] that is released in the blood and plays several roles in viral replication and immune evasion [8, 9]. This unique feature enables the detection of NS1, making it a biomarker of dengue during acute infection. High levels of NS1 protein can be detected since the first symptoms up to 9 days of disease [10]. Alcon et al. reported NS1 levels in human serum from 0.04 to 2 µg/ml in the first and 0.01 to 2 µg/ml in the second infection [11].
Nowadays, the clinical tests are based on the detection of NS1 using IgM and IgG antibodies in enzyme-linked immunosorbent assay (ELISA), a current standard technique or nucleic acid amplification tests for RNA [8]. All these laboratory methodologies are efficient; however, they require expensive equipment and specific and technical expertise [9, 12].
Sensitive platforms using electrochemical techniques for biosensors have made great progress, mainly due to the efforts of different research fields. Electrochemical, and especially potentiometric, biosensors are one of the most promising classes with relatively simple devices, low fabrication costs, and easy miniaturization processes [9]. Electrochemical impedance spectroscopy (EIS) is widely used to investigate modified surfaces capable of detecting biomarkers. However, the need for a redox probe in solution to obtain the data reduces the possible applications. Cecchetto et al. reported a sensitive electrochemical capacitance spectroscopy (ECS) technique for dengue detection through antibodies [12]. One other promising possibility is the use of specific and artificial DNA sequences (aptamers) to interact with the biomarker [13]. Compared with immunoassay-based methods, which are strongly dependent on antibody quality, aptamers exhibit a high specificity, affinity, excellent stability, easy modification, and low cost to develop the biosensing platform [14].
In a previous study [15], the focus was investigating the aptamer interaction with two different NS1 serotypes using impedimetric detection. However, in this method, it was necessary to add a redox molecule to the solution to verify and quantify the interaction. In this study, gold electrodes were modified with DNA aptamers to detect NS1 protein using ECS. This device was based on a self-assembled monolayer (SAM) where the aptamers were co-immobilized with 6-(ferrocenyl)hexanethiol (FCH) and 6-mercapto-1-hexanol (MCH). The probe surface density was optimized, and the biosensor performance was tested in phosphate-buffered saline (PBS) and undiluted human serum.
Experimental
Materials
The Serotype 4 NS1 Protein (Abcam, ab181957) was used as a target. The DNA aptamer sequence (5′-HS(CH2)6–TTTTT ACTAGGTTGCAGGGGACTGCTCGGGATTGCGGATCAACCTAGTTGCTTCTCTCGTATGAT–3′), Trizma base (C4H11NO3, ≥ 99%), magnesium chloride (MgCl2, ≥ 98%), octaethylene glycol monododecyl ether (C12E8, ≥ 97%), 6-(ferrocenyl)hexanethiol (C16H22FeS, ≥ 97%), 6-mercapto-1-hexanol (HS(CH2)6OH, ≥ 97%), bovine serum albumin (BSA), sodium phosphate dibasic (Na2HPO4, ≥ 99%), potassium phosphate monobasic (KH2PO4, ≥ 99%), and ethylenediaminetetraacetic acid (C10H16N2O8, ≥ 99%) were purchased from Sigma-Aldrich. Sulfuric acid (H2SO4, ≥ 95%) and sodium chloride (NaCl, ≥ 99%) were purchased from Synth; sodium hydroxide (NaOH, ≥ 97%) was purchased from Dinamica and potassium chloride (KCl, ≥ 99%) was purchased from Cinetica. Ultra-pure water (18.2 MΩ.cm, Merck Millipore Direct-Q 5UV) was used to prepare all aqueous solutions. The Biopak Polisher filter (Merck Millipore, CDUFB-001) was used in Millipore equipment to remove all nuclease in ultra-pure water. Commercial human serum was purchased from Sigma-Aldrich (human male AB plasma, USA origin, sterile-filtered) and stored in the freezer (− 20 °C). All the chemicals used in this study were analytical or commercial grade.
Electrode preparation
All procedures were developed by your group and published in previous works [15]. Briefly, gold electrodes with a 2 mm diameter (Metrohm) were desorbed using cyclic voltammetry in NaOH (0.5 M) from − 2.0 to − 0.8 mV (vs. Ag/AgCl) for 30 cycles. Thereafter, the surface was polished with different alumina sizes (1 and 0.3 µm), followed by sonication in water for 5 min between each polish. The electrochemical cleaning was performed in H2SO4 (0.5 M) by scanning the potential from − 0.5 to 1.1 V (vs. Ag/AgCl) for 25 cycles [16]. The effective surface was calculated by the reduction peak area and used to normalize all data [17, 18]. Clean electrodes were dried with N2.
Electrode functionalization
DNA aptamer was incubated at 94 °C in a water bath for 10 min and immediately diluted to 1 µg/mL in Selex solution (20 mM Trizma pH 7.5 + 150 mM NaCl + 1 mM MgCl2 + 2.7 mM KCl + 0.005% C12E8) at room temperature (24 °C) for 40 min [13, 15]. The Selex solution with aptamer was mixed with 1 mM 6-(ferrocenyl)hexanethiol (FCH) (10% ethanol) in different molar fractions. The fractions are presented in this work as (Apt / (total thiol)), where total thiol corresponds to the sum between aptamer and FHC. The electrodes were co-immobilized with aptamer and FHC in 150 µl of Selex solution for 18 h, in a humidity chamber, at 4 °C [19].
Modified gold electrodes were rinsed with three wash solutions (200 mM phosphate buffer (PB); 10 mM PB + 100 mM ethylenediaminetetraacetic acid (EDTA); 10 mM PB) for 5 min each. Following this procedure, the electrodes were backfilled with 1 mM 6-mercapto-1-hexanol (MCH, 10% ethanol) for 1 h to block the remaining spaces on the surface. The electrodes were rinsed with ultra-pure water (Milli-Q) and immediately placed in PBS (137 mM NaCl + 2.7 mM KCl + 1 mM Na2HPO4 + 0.18 mM KH2PO4, pH 7.4) for 2 h to stabilize [16]. Finally, the gold electrodes were exposed to BSA (0.1%) or serum for 30 min. Figure 1a presents an illustration of SAM architecture. The capacitance responses from blocked electrodes with BSA, or serum, were used as a baseline for all NS1 responses. Figure 1b exhibits the capacitive response after the BSA blocking and interaction with 1 µg/mL NS1 protein.
Schematic illustration of self-assemble monolayer in a. The illustration is out of scale and does not represent the real aptamer folding. In b, the capacitive signal after the BSA blocking. The signal decreases upon the interaction with 1 µg/mL of NS1 protein. In detail, the decrease in capacitance curve
Measurement and apparatus
All measurements were obtained using the potentiostat (Metrohm/Autolab PGSTAT302) with a three-electrode cell configuration: Ag/AgCl (FACTE) as the reference electrode, platinum wire (Metrohm) as the counter electrode, and gold electrode as the working electrode. All procedures were carried out in a 15-ml electrochemical cell with PBS buffer. The ECE measurement was performed in the 100-kHz to 100-mHz frequency range with a 10 mV a.c. potential difference superimposed on a d.c. bias of the open circuit potential (OCP) configuration in PBS buffer. Autolab software (Nova 2.1) was used. The OCP measurement can be used to detect the potential variation due to the changes in biolayer capacitance [19]. To analyze the charge on the surface, the OCP values obtained for SAM/BSA response were fixed during all procedures for each electrode. All data were originally obtained as complex impedance signals (Z*(\(\omega )\)), Eq. 1. The capacitance complex signals (C*(\(\omega\))) were obtained by deriving the last equation in two new equations (Eqs. 2 and 3):
where C′ is the real capacitance; C″ is the imaginary capacitance [20]. These data generated a capacitive spectrum (Fig. 1b).
The biosensor surface was characterized via atomic force microscopy (AFM) using a Shimadzu SPM-9600 Scanning Probe Microscopy (Shimadzu Corporation, Japan). The analysis was obtained in air at 25 °C using the same gold electrode.
Results and discussion
Electrochemical capacitance spectroscopy analysis
The EIS is composed of two processes: Faradaic and non-Faradaic. The Faradaic process needs a redox specie (e.g., the redox couple [Fe(CN)6]4−/[Fe(CN)6]3−) to investigate the interactions on the electrode surface [16]. The non-Faradaic measurements do not require any external redox probe, indicating the existence of dielectric (associated with a double layer) contribution and making this technique able to analyze small changes in the interface charge [21]. In this case, the technique uses a redox probe (FCH) co-immobilized with the biological receptive layer. The capacitance values are related to this molecule’s coverage and concentration. The interaction between the aptasensor and target changes the charge over the surface, limits the redox process, and reduces the capacitive response [12].
To analyze the capacitive data, it is important to find the highest phase angle by plotting the dose-dependent response from the capacitive signal change over frequency [22]. The phase angle is presented in Fig. 2a and indicates the maximum capacitive response at ~ 1.7 Hz, which corresponds to 86°. Fixing the analysis in 1.7 Hz, Fig. 2b shows the real capacitance (C′) reduction when the NS1 concentration increases from 1 pg/mL to 1 µg/mL, indicating a successful binding between protein and aptamer. Figure 2c exhibits the alteration in dielectric properties on the biolayer capacitance for different protein concentrations. The interface SAM/BSA indicates 1.19 µF as capacitance response, and this value decreases to 1.15 µF with the addition of 1 µg/mL NS1 protein, which corresponds to 40-nF variance in real capacitance (− ΔC′).
Surface functionalization
The surface modification for ECS measurements was tested with two different protocols. First, the surface was co-immobilized with DNA aptamer and FCH. After 18 h, the electrodes were backfilled with MCH. This result is presented in Fig. 3 and corresponds to aptamer + FCH/MCH. In the second protocol, aptamer and MCH were co-immobilization and backfilled with FCH (aptamer + MCH/FCH).
Figure 3 shows the change in real capacitance [− ΔC′ = (CNS1 − CSAM)] over NS1 concentrations from 10 pg/mL to 1 \(\upmu\) g/mL. When the surface is co-immobilized with aptamer + FCH, the NS1 increase changes the capacitance response. However, the aptamer + MCH co-immobilization with FCH just added in the backfilling does not show any increase in capacitance response. This result is related to the presence of FCH on the SAM; once it takes more time to interact with the gold surface and organize as part of the SAM [23], the backfilling is not appropriate.
When FCH is correctly immobilized on the gold surface, it can accumulate charge in the interface, increasing the capacitive value and generating a high electron transfer rate between interface and surfac [24, 25], which can be observed with the change in − ΔC′ when NS1 interacts with aptamer.
Surface characterization and protein interaction were also characterized using atomic force microscopy (AFM). The images are shown in Figure S1 (Supplementary information), where the clean bare electrode (Figure S1(a)) exhibited an 83.53-nm peak. After surface immobilization, backfilling, and BSA blocking, the peak value increased to 145.10 nm, indicating that the immobilization of biomolecules on the gold surface increased the surface roughness due to the different sizes of the molecules (Figure S1(b)). After 1 µg/mL NS1 interaction, the peak value decreased to 101.34 nm. This change indicated that the protein interaction on the surface occupies the remaining spaces and brings the aptamers close to the electrode surface, resulting in a smoother surface (Figure S1 (c)).
Surface density
Auletta et al. report that the proportion between FCH and aptamers influences the interaction between target and probe [26]. Electrodes were exposed to different molar fractions of FCH and aptamers (1:10, 1:50, 1: 100, 1: 150, 1: 200) in immobilization solution and tested with 100 pg/mL NS1 diluted in buffer solution (PBS + BSA (0.1%), pH 7.4). Figure 4 shows the influence of molar fraction in capacitance response as an important step for the biosensor performance, once FCH concentration affects the SAM organization. The surface coverage does not need a large amount of aptamer (1:10) or FCH (1:200) because the SAM organization depends on the alkanethiol chain and van der Walls interactions, making an intermolecular film in the gold surface [27]. The ratio 1:10 showed the lowest capacitance signal (2 nF), which indicates an excess of aptamer and high repulsion between DNA oligonucleotide aptamer chains, generating an unstable monolayer. On the other hand, the ratio 1:200 presented a low capacitance signal (4 nF) due to the low amount of aptamer. The most stable biolayer was performed by 1: 150 molar fraction (with 11 nF) and was used for biosensor development.
Analytical curve and negative control
The optimized ratio was used to test the system and obtain a calibration plot. The performance of this biosensing platform was tested from 10 pg to 1 µg/mL in buffer (Fig. 5a) and commercial undiluted human serum (Fig. 5b). For each measurement, the SAM was incubated with 100 µl of NS1 concentrations for 30 min, followed by washing with PBS to remove unbound NS1.
Analytical curve for NS1 diluted with a linear response in a buffer; b commercial undiluted human serum; c change in real capacitance (− ΔC′) for each NS1 concentration in buffer (open black square) and undiluted serum (open red circle). Error bars show the standard deviation from at least three individual electrodes. d Negative control shows the change in real capacitance (− ΔC′) for 1 ng/mL of NS1 and envelope protein (protein E) in serum
Figure 5c shows the calibration plot as − ΔC′ increases for NS1 concentrations in buffer and undiluted serum. The platform has a linear behavior from 10 pg/mL to 1 µg/mL (buffer) and from 10 pg/mL to 100 ng/mL (undiluted serum). Serum has a high amount of protein that can saturate an electrode surface, reducing the linear range.
The limit of detection (LOD) was calculated as 3.3 × σ/b, where σ represents standard deviation, and b represents the slope of the analytical curve fitting on a linear range [28]. In buffer, the biosensor presented a LOD of 30.9 fg/mL with 3-nF sensitivity per decade. In undiluted serum, the platform reached 41.8 fg/mL and a sensitivity of 4 nF per decade. The relative standard deviation (RSD) was calculated from both calibration curves (0.09% and 0.1% from PBS and serum, respectively). These results exhibited a high sensibility in comparison with other techniques reported in the literature. Table S1 (supplementary material) compares the results from the last 5 years of different platforms to detect NS1 in the clinical range for first (0.04 to 2 µg/mL) and second (0.01 to 2 µg/mL) infection.
The negative control was performed with 1 ng/mL E protein in undiluted serum. Envelope protein (E protein) is a dengue protein expressed in infected cells and is responsible for virus protection and target cell binding [29]. Figure 5d shows the difference between the NS1 response with 14 nF in comparison to the E protein (8 nF) with a 43% decrease in the signal response. This difference validates the selectivity characteristic of this aptamer to detect Dengue NS1 protein.
It is crucial to acknowledge the current limitations of this platform, including its non-disposable electrodes, and the need for improved NS1 detection in real human samples. All the measurements were performed with at least six different electrodes and performed at two different times. The results present a 5% difference between batches. The aptamer platform storage lifetime was previously tested using other sequences, in the same conditions, and presented a 40% signal reduction after 2 weeks. Despite these limitations, the proposed method for capacitive biosensors enables dengue and other disease detection using a label-free system.
Conclusion
A new platform that selectively quantifies NS1 protein using DNA aptamer on the gold electrode was developed. As a highlight, it was demonstrated that electrochemical capacitive spectroscopy, using a simple approach with FCH on the SAM, was able to detect NS1 in PBS and in serum biological matrix presenting a linear detection range from 0.01 to 1000 ng/mL in buffer and 0.01 to 100 ng/mL in undiluted serum. The biosensor achieved low limits of detection of 30.9 fg/mL and 41.8 fg/mL in buffer and undiluted serum, respectively. The main advantage of this platform is that there is no need to add a redox molecule to the solution, which is particularly useful to reduce patient sample manipulation, increasing applications. Moreover, the ECS platform exhibited a rapid response time, easy operation, and cost-effectiveness.
Data availability
The data that support the findings of this article are available from the authors upon request.
References
Ten health issues WHO will tackle this year. https://www.who.int/news-room/feature-stories/ten-threats-to-global-health-in-2019. Accessed 16 Apr 2020
Redoni M, Yacoub S, Rivino L et al (2020) Dengue: status of current and under-development vaccines. Rev Med Virol e2101. https://doi.org/10.1002/rmv.2101
Bhatt S, Gething PW, Brady OJ et al (2013) The global distribution and burden of dengue. Nature 496:504–507. https://doi.org/10.1038/nature12060
Guzman MG, Halstead SB, Artsob H et al (2010) Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–S16. https://doi.org/10.1038/nrmicro2460
de GóesCavalcanti LP, Freitas ARR, Brasil P, da Cunha RV (2017) Surveillance of deaths caused by arboviruses in Brazil: from dengue to chikungunya. Mem Inst Oswaldo Cruz 112:583–585. https://doi.org/10.1590/0074-02760160537
Atualização Epidemiológica Dengue, chikungunya e Zika -25 de janeiro de 2023 - OPAS/OMS | Organização Pan-Americana da Saúde. https://www.paho.org/pt/documentos/atualizacao-epidemiologica-dengue-chikungunya-e-zika-25-janeiro-2023. Accessed 7 Jul 2023
Halstead SB, Russell PK, Brandt WE (2020) NS1, Dengue’s dagger. J Infect Dis 221:857–860. https://doi.org/10.1093/infdis/jiz083
Foley DA, Yeoh DK, Karapanagiotidis T et al (2020) Fever in the returned traveller: the utility of the Platelia Dengue NS1 antigen enzyme immunoassay for the diagnosis of dengue in a non-endemic setting. Pathology (Phila) 52:370–372. https://doi.org/10.1016/j.pathol.2020.01.438
Villalonga A, Pérez-Calabuig AM, Villalonga R (2020) Electrochemical biosensors based on nucleic acid aptamers. Anal Bioanal Chem 412:55–72. https://doi.org/10.1007/s00216-019-02226-x
Teoh B-T, Sam S-S, Tan K-K et al (2016) The Use of NS1 Rapid diagnostic test and qRT-PCR to complement IgM ELISA for improved dengue diagnosis from single specimen. Sci Rep 6:1–8. https://doi.org/10.1038/srep27663
Alcon S, Talarmin A, Debruyne M et al (2002) Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J Clin Microbiol 40:376–381. https://doi.org/10.1128/JCM.40.02.376-381.2002
Cecchetto J, Fernandes FCB, Lopes R, Bueno PR (2017) The capacitive sensing of NS1 Flavivirus biomarker. Biosens Bioelectron 87:949–956. https://doi.org/10.1016/j.bios.2016.08.097
Lee KH, Zeng H (2017) Aptamer-based ELISA assay for highly specific and sensitive detection of Zika NS1 protein. Anal Chem 89:12743–12748. https://doi.org/10.1021/acs.analchem.7b02862
Jia M, Sha J, Li Z et al (2020) High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem 317:126459. https://doi.org/10.1016/j.foodchem.2020.126459
Bachour Junior B, Batistuti MR, Sanches Pereira A et al (2021) Electrochemical aptasensor for NS1 detection: towards a fast dengue biosensor. Talanta 233:122527. https://doi.org/10.1016/j.talanta.2021.122527
Keighley SD, Li P, Estrela P, Migliorato P (2008) Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens Bioelectron 23:1291–1297. https://doi.org/10.1016/j.bios.2007.11.012
Benites TA, Ribeiro WC, Góes MS et al (2014) Effects of surface roughness on properties of passivation of self-assembled organic monolayers. Quím Nova 37:1533–1537. https://doi.org/10.5935/0100-4042.20140241
Trasatti S, Petrii OA (1991) Real surface area measurements in electrochemistry. Pure Appl Chem 63:711–734. https://doi.org/10.1351/pac199163050711
Wong LCC, Jolly P, Estrela P (2018) Development of a sensitive multiplexed open circuit potential system for the detection of prostate cancer biomarkers. BioNanoScience 8:701–706. https://doi.org/10.1007/s12668-017-0408-0
Fernandes FCB, Góes MS, Davis JJ, Bueno PR (2013) Label free redox capacitive biosensing. Biosens Bioelectron 50:437–440. https://doi.org/10.1016/j.bios.2013.06.043
Wang L, Veselinovic M, Yang L et al (2017) A sensitive DNA capacitive biosensor using interdigitated electrodes. Biosens Bioelectron 87:646–653. https://doi.org/10.1016/j.bios.2016.09.006
Singh NK, Arya SK, Estrela P, Goswami P (2018) Capacitive malaria aptasensor using Plasmodium falciparum glutamate dehydrogenase as target antigen in undiluted human serum. Biosens Bioelectron 117:246–252. https://doi.org/10.1016/j.bios.2018.06.022
Ulman A (1996) Formation and structure of self-assembled monolayers. Chem Rev 96:1533–1554. https://doi.org/10.1021/cr9502357
Marchante E, Crivillers N, Buhl M et al (2015) An electrically driven and readable molecular monolayer switch based on a solid electrolyte. https://doi.org/10.1002/ange.201508449
Rivera IM, Cabrera CR (1993) Mass measurements of ferrocene adsorption at gold films with the electrochemical quartz crystal microbalance. J Electrochem Soc 140:L36–L38. https://doi.org/10.1149/1.2056183
Auletta T, van Veggel FCJM, Reinhoudt DN (2002) Self-assembled monolayers on gold of ferrocene-terminated thiols and hydroxyalkanethiols. Langmuir 18:1288–1293. https://doi.org/10.1021/la011474u
Kleinjung F, Klussmann S, Erdmann VA et al (1998) High-affinity RNA as a recognition element in a biosensor. Anal Chem 70:328–331. https://doi.org/10.1021/ac9706483
Armbruster DA, Pry T (2008) Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 29:S49–S52
Chen Y, Maguire T, Hileman RE et al (1997) Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3:866–871. https://doi.org/10.1038/nm0897-866
Funding
BBJ was financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Brasil (CAPES—Finance Code 001). MRBS received funding from FAPESP (2013/26133–7) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (151658/2022–6). MM received funding from FAPESP (2017/24201–6) and CNPq (308713/2018–4 and 310405/2021–1).
Author information
Authors and Affiliations
Contributions
BBJ carried out the experimental work under the supervision of MRB and MM. MRB and BBJ wrote the paper with contributions from MM. All authors checked and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bachour Junior, B., Batistuti Sawazaki, M.R. & Mulato, M. Electrochemical capacitive dengue aptasensor using NS1 in undiluted human serum. Microchim Acta 191, 72 (2024). https://doi.org/10.1007/s00604-023-06141-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-06141-y