Abstract
A convenient fluorometric method was developed for specific determination of DNA based on peptide nuclei acid (PNA)–regulated fluorescence resonance energy transfer (FRET) between carbon dots (CDs) and gold nanoparticles (AuNPs). In this system, CDs that display lake blue fluorescence with excitation/emission maxima at 345/445 nm were used as fluorometric reporter, while AuNPs were used as fluorescence nanoquencher. A neutral PNA probe, which is designed to recognize the target DNA, was used as a coagulant to control the dispersion and aggregation of AuNPs. Without DNA, PNA can induce immediate AuNP aggregation, thus leading to the recovery of the FRET-quenched fluorescence emission of CDs. However, the addition of the complementary target DNA can protect AuNPs from being aggregated due to the formation of DNA/PNA complexes, which subsequently produces a high fluorescence quenching efficiency of CDs by dispersed AuNPs. Under optimized conditions, quantitative evaluation of DNA was achieved in a linear range of 5–100 nM with a detection limit of 0.21 nM. This method exhibited an excellent specificity towards fully matched DNA. In addition, the application of this assay for sensitive determination of DNA in cell lysate demonstrates its potential for bioanalysis and biodetection.

A simple fluorometric biosensor for specific detection of DNA was developed based on peptide nuclei acid (PNA)–regulated fluorescence resonance energy transfer (FRET) between carbon dots (CDs) and gold nanoparticles (AuNPs).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the genetic basis of disease is gradually uncovered, the medical diagnosis and treatment based on pathogenic gene detection is becoming increasingly important [1, 2]. While significant progress has been made with polymerase chain reaction (PCR) and next-generation sequencing (NGS), the most common method to detect and quantify nucleic acid, it is still very difficult to popularize them in most public health settings due to the high testing cost, its being time-consuming, and the requirement of specialized instrument and laboratory environment. Thus, the development of rapid, economical, and accurate method for specific DNA detection and quantification is still in urgent need. In the past decades, the design of fluorescent biosensors for specific DNA has attracted considerable attention owing to their inherent advantages including high sensitivity, ease of operation, and in situ imaging properties [3,4,5,6,7]. It is worth noting that fluorescence resonance energy transfer (FRET) or quenching mechanism-based DNA assays are one of the most popular strategies, such as famous molecular beacon hairpin structures [8, 9]. They usually comprise of a fluorophore and a quencher molecule respectively attached to two ends of a DNA probe strand. In the absence of the target molecule, the fluorophore and quencher molecules are in close proximity and fluorescence is quenched. Upon hybridization with the target molecule, the quencher molecule becomes physically separated from the fluorophore molecule and fluorescence is recovered. However, although these fluorescent assays have been extensively developed in a series of applications [10,11,12], they still suffer from several drawbacks, such as careful selection of fluorescence-quenching pairs, need of probe labeling, and the shortcomings from used fluorescent organic dyes which contain sophisticated synthetic procedures, rapid photobleaching, narrow excitation spectra, broad emission band, small Stokes shifts, high cost, and a short fluorescence lifetime [13].
Carbon dots (CDs), a novel fluorescent carbon nanomaterial with a size below 10 nm, are gradually becoming a rising star in the research of fluorescence analysis [14]. Compared with organic dyes and traditional quantum dots, CDs are proudly characterized by easy preparation, tunable excitation and emission, high photostability, inexpensive raw materials, and low cytotoxicity [15, 16]. These superior and unique properties make CDs ideal probes in fluorescent biosensors [17,18,19]. On the other hand, gold nanoparticles (AuNPs) are ideal fluorescence quenchers due to their high extinction coefficients and a wide absorption spectral range, which can overlap with the emission of many fluorophores [20,21,22]. In the past two decades, FRET based on CD/AuNP pair has been widely utilized for sensing of small molecules [23,24,25,26,27,28,29,30,31] and protein [32, 33]. In these studies, CDs acted as a highly efficient fluorescent donor and AuNPs acted as an excellent acceptor to substitute traditional organic dyes and quenchers. The FRET distance between quantum dots (QDs) and AuNPs was modulated by the dispersion/aggregation state of AuNPs or the competitive replacement of CDs on the surface of AuNPs. Nevertheless, the determination of DNA using these methods has not been sufficiently explored. To the best of our knowledge, there are only two studies reporting CD-AuNP systems for DNA sensing applications [34, 35]. In their designs, DNA probes need to be labeled and the adoptive CDs or AuNPs need to be modified and separated, which make them cumbersome, expensive, and time-consuming. Although the CD-AuNP/graphene oxide (GO) system is highly sensitive, it is hard to distinguish single-base mismatched targets from fully-matched ones [35], which also limits their applications in pathogenic gene detection, because many human diseases are caused by single-base mutations [36,37,38].
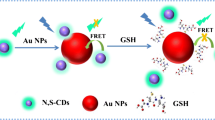
Considering the flexibility, simplicity, and high specificity of DNA detection process, we propose a peptide nucleic acid (PNA)–regulated FRET method based on CDs and AuNPs for specific DNA determination (Scheme 1). PNA are synthetic DNA/RNA analogues in which the entire sugar phosphate backbone is replaced by an uncharged polyamide backbone [39]. Distinct backbone properties, especially charge neutrality, not only endow PNA with outstanding hybridization properties such as higher binding affinity and better specificity but also bring an exceptional interaction with nanomaterials [40,41,42]. It was reported that PNA can induce immediate aggregation of citrate ion-coated AuNPs, while PNA-DNA complexes can effectively keep the particles dispersed [40, 41]. Therefore, it is reasonable to infer that the combination of CD/AuNP pair and PNA probe would have a good application prospect in the establishment of more convenient, cost-effective, specific, and sensitive methods for DNA determination, which avoids modification and separation steps, and this would potentially broaden the applicability of CD/AuNP pair–based fluorometric method.
Experimental section
Materials and reagents
All of the 9-fluorenylmethoxycarbonyl (Fmoc)-PNA monomers were purchased from PANAGENE Inc. (Daejeon, Korea, http://www.panagene.com). Methyl-benzhydrylamine (MBHA) resin was purchased from CSBio Ltd. (USA, https://www.csbio.com/offices.html). DNA oligonucleotides were synthesized and purified by Jie Li Biology Inc. (Shanghai, China, http://www.genebioseq.com/). All PNA and DNA sequences were listed in Table S1. Malic acid, urea, ethyl acetate, petroleum ether, chloroauric acid, sodium citrate, ethylenediaminetetraacetic acid disodium salt (EDTA), acetic anhydride (Ac2O), dichloromethane (DCM), N,N-diisopropylethylamine (DIEA), N,N-dimethylformamide (DMF), Kaiser reagent, N-methyl-2-pyrrolidone(NMP), trifluoroacetic acid (TFA), triisopropylsilane, tris(hydroxymethyl)aminomethane, anhydrous diethyl ether, potassium chloride, acetonitrile, O-benzotriazole-tetramethylurea hexafluorophosphate (HBTU), piperidine, and pyridine were purchased from Aladdin (Shanghai, China, https://www.aladdin-e.com/). HAuCl4·3H2O (99.9%) and trisodium citrate dihydrate (99.5%) were purchased from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China, http://en.reagent.com.cn/). HeLa cell was purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China, http://www.cellbank.org.cn/). The RPMI 1640 medium and PBS were purchased from Thermo Scientific (USA, https://www.thermofisher.com/). RIPA lysis buffer was purchased from Beyotime Biotech (China, https://www.beyotime.com/index.htm). Ultrapure water (18 MΩ·cm.) was prepared from Millipore (Merck) Milli-Q water system. All experiments were performed in Tris-HCl buffer (2 mM Tris-HCl, 5 mM NaCl, pH 7.4) unless otherwise stated.
Apparatus
Fluorescence spectroscopy measurements were performed on a LS55 fluorescence spectrophotometer (PerkinElmer, USA, http://www.perkinelmer.com.cn/). The ultraviolet-visible (UV-vis) spectra was obtained by a Cary 300 UV-vis spectrophotometer (DeNovix, USA, http://www.denovix.com/). Transmission electron microscope (TEM) images were collected from a JEM2100 transmission electron microscope (JEOL, Japan, https://www.jeol.co.jp/). Fourier transform infrared spectra (FT-IR) were recorded on a Nicolet is50 Fourier transform infrared spectrometer (ThermoFisher, USA, https://www.thermofisher.com/). PNA was purified using a 1260 high performance liquid chromatograph (Agilent, USA, https://www.agilent.com/about/). The molecular weight of the PNA was determined on a 4600 time-of-flight mass spectrometer (AB Sciex, USA, https://sciex.com.cn/).
PNA synthesis
PNA was manually synthesized by the solid-phase peptide synthesis protocol as we reported previously [43]. The crude PNA was cut off from the resin in a cleavage cocktail [43] and precipitated out in plenty of anhydrous diethyl ether. Then PNA precipitate was dried by carefully passing a stream of dry N2 and dissolved in ultrapure water. PNA was purified by a reverse-phase column (Agilent Eclipse XDB-C18) and characterized by mass analysis (see Fig. S1). The concentration of PNA was calculated by the absorbance at 260 nm and the extinction coefficient, which was estimated based on the sum of the extinction coefficients of component bases. At last, a stock solution of PNA (100 μM) is prepared in high-purity water for use.
Preparation of CDs and AuNPs
CDs were synthesized by the solvothermal treatment of malic acid and urea. Briefly, 10 mM malic acid (1390.9 mg) and 10 mM urea (600.6 mg) were fully mixed in a 10 mL DMF solution. Then the mixture was poured into a 100-mL autoclave and placed in an oven at 200 °C for 15 h. The resulting dark brown solution was centrifuged at 6481×g (10,000 rpm) for 10 min, and the supernatant was collected, followed by dialysis using a 1 kDa cutoff membrane for 1 week. The CD solution was next freeze-dried to powder before subsequent use. The concentration of CDs used in this experiment is 5 μg mL−1. Citrate-stabilized AuNPs with a diameter of about 13 nm were prepared using a standard protocol [41, 44], and details are given in the Electronic Supporting Material.
DNA assay
For the determination of target DNA, different concentrations of DNA were first annealed with a specific concentration (0.8 μM) of PNA in Tris-HCl buffer at room temperature. After 10 min, 50 μL of 5 nM AuNPs and 10 μL of CDs were added to the sample solution. Then the mixture was incubated at room temperature for 1 h, and the change in fluorescence intensity was analyzed by a fluorescence spectrophotometer at an excitation wavelength of 345 nm. For the specificity analysis, a similar protocol was performed except that the mismatched DNA was used instead of the target DNA.
Preparation of cell lysate
HeLa cells were trypsinized, collected by centrifugation, and washed with 0.1 M PBS. Then the cells were centrifuged again at 373×g (2400 rpm) for 5 min at 4 °C to remove the supernatant. The collected cells were suspended in RIPA lysis buffer at a concentration of 5.5 × 106 cell∙mL−1 [41], and then the resultant suspension was incubated on ice for 40 min. Finally, the lysis products were centrifuged at 9333×g (12,000 rpm) for 30 min at 4 °C. The supernatant was collected and stored in − 20 °C for use. In this experiment, a 10% HeLa cell lysate was used for target DNA assay.
Results and discussion
Synthesis and characterization of CDs and AuNPs
Novel N-doped CDs were synthesized by the solvothermal method using malic acid and urea as carbon resources. As displayed in Fig. 1a, TEM image shows that CDs are well dispersed with nearly spherical shape, and the average diameter calculated from about 100 particles is 3.5 nm (Fig. S2). Lattice fringes of CDs are clearly observed, and the inset shows the lattice spacing distance of about 0.21 nm, which is similar to other CDs reported previously [26, 29]. The functional groups on the surface of the synthesized CDs were presented using FT-IR spectra. As described in Fig. 1b, the characteristic absorption peaks appear at 1100 cm−1, 1195 cm−1, 1405 cm−1, 1715 cm−1, 3170 cm−1, and 3340 cm−1, and they are usually caused by the stretching vibrations of C–O, C–N, C=C, C=O, N–H, and O–H, respectively. Especially the presence of strong absorption bands of 1715 cm−1, 3170 cm−1, and 3340 cm−1 suggests that there were plenty of carboxyl groups and amino groups on the surface of CDs. Among them, the existence of amino groups contributes to a better fluorescence quenching effect because FRET distance is shortened through the Au-N interaction between AuNPs and CDs.
Citrate-stabilized AuNPs were prepared by the reduction of HAuCl4 with sodium citrate and characterized by TEM and UV-vis absorption spectra. Figure 1c and d show that AuNPs are uniform and monodisperse, with an average diameter of 13 nm and a strong characteristic surface plasmon resonance (SPR) absorption peak at 520 nm. These results indicate the successful synthesis of gold nanoparticles.
Then the optical properties of CDs were investigated using both UV-vis absorption and fluorescence spectroscopy (Fig. 2). It is found that CDs possessed two obvious absorption peaks at 287 nm and 350 nm (spectrum UV-vis), which ascribes to the large amount of π-π* transition of CDs and the n-π* transition of C=O bond, respectively [26, 32]. CD solution is amber, clear, and transparent under sunlight and exhibited bright lake blue luminescence under UV light (inset in Fig. 2). This demonstrates that the CDs have an excellent fluorescence property. The photoluminescence (PL) emission spectra of CDs display an obvious excitation-dependent feature, the emission peak gradually shifts to longer wavelength as the excitation wavelength increase (data not shown), with an excitation maximum at 345 nm (spectrum Ex) and an emission maximum at 445 nm (spectrum Em). It can be seen that the emission band of CDs (spectrum Em) overlaps with the absorption band of dispersed AuNPs (spectrum Dispersed). This implies that the CDs and AuNPs can function as a donor-acceptor pair for a FRET system. Zeta potential measurements show that CDs are positively charged (+ 8.85 mV) and AuNPs are negatively charged (− 36.8 mV) (Fig. S3). The electrostatic interaction between positively charged CDs and negatively charged AuNPs can shorten the distance between CDs and AuNPs, which is beneficial to the occurrence of FRET.
UV-vis absorption spectra (UV-vis), fluorescence excitation (Ex), and emission spectra (Em) of CDs; UV-vis absorption spectra of PNA-DNA heteroduplex-coated AuNPs (Dispersed, similar to original AuNPs) and PNA-coated AuNPs (Aggregated). The inset shows a photograph of CDs under sunlight (a) and UV light (b)
Assay mechanism
The principle of the PNA-regulated FRET method for DNA detection is illustrated in Scheme 1. A 12-mer PNA probe is used to selectively recognize the complementary target DNA and to regulate the aggregation and dispersion of AuNPs. PNA was designed and synthesized using standard Fmoc chemistry as described previously [43]. In a typical sensing process, the probe PNA is first incubated with target DNA followed by the addition of AuNPs and CDs. It has been reported that dispersed AuNPs can effectively quench the fluorescence of CDs via FRET [32]. So in this system, if no target DNA exists, free PNA molecules in solution will induce particle aggregation immediately and robustly. This is result from the removal of charge repulsion between the neutral PNA and the citrate anion-coated AuNPs [40]. The PNA probe induced particle aggregation was verified by both TEM (Fig. S4) and a characteristic solution color change from red to blue. Then, the decrease in the absorbance at the characteristic SPR absorption peak (521 nm) and the appearance of absorption peak at longer wavelength (644 nm) further confirm the aggregation of AuNPs (Fig. 2, spectrum Aggregated). As a result, the overlap between the SPR absorption peak of AuNPs and the emission band of CDs is reduced, and the fluorescence quenching efficiency of AuNPs towards CDs is decreased. Therefore, the fluorescence of CDs is turned on. However, in the presence of the complementary target DNA, PNA-DNA complexes can protect AuNPs from salt-induced aggregation [40], which results in an effective quenching of the fluorescence of CDs (Fig. 2, spectrum Dispersed). Based on this principle, the reduction of CD emission intensity, that is, the fluorescence quenching extent, can be used to quantify the amount of target DNA.
Verification of assay mechanism
In consideration of a FRET-based assay mechanism, a series of fluorescent experiments were first carried out to confirm the feasibility of the method. As depicted in Fig. 3a, CDs alone exhibit a strong fluorescence peak at 445 nm (spectrum a). When AuNPs was added to the solution of CDs, a dramatic fluorescence quenching is observed (spectrum b) due to the FRET between CDs and AuNPs. However, after the addition of PNA to the mixture of AuNPs and CDs, the aggregation of AuNPs occurs and the fluorescence intensity emitted by CDs is largely restored (spectrum c). The characteristic SPR peak of the CD/AuNP solution at 521 nm is red-shifted to 627 nm (Fig. 3b). These results suggest that the adsorption of the electroneutral PNA probe lead to the aggregation of AuNPs even in the presence of CDs, which reduce the overlap between CD emission spectrum and AuNP SPR absorption band. The interaction region between CDs and agglomerated AuNPs becomes smaller than that of free AuNPs. These results ultimately bring about a lower FRET efficiency. Whereas when a fully matched target DNA was added, the fluorescence quenching efficiency of CDs is greatly enhanced (Fig. 3a, spectrum d) and no obvious change in the absorption spectrum is observed (Fig. 3b, spectrum d). This indicates a higher FRET efficiency between CDs and the PNA-DNA complex stabilized AuNPs. According to the mechanism of FRET, the more overlap between the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor, the better quenching effect. Thus, the dispersed AuNPs displayed a stronger fluorescence quenching ability than the aggregated ones. We notice a reduced fluorescence quenching (Fig. 3a, spectrum d) for that of the PNA-DNA complex–treated AuNPs compared with that of AuNPs alone (spectrum b), which may be caused by the adsorption of PNA-DNA complex onto AuNP surface. To exclude the possibility that the reduced fluorescence quenching efficiency is caused by the adsorption of the ssDNA onto AuNPs, control experiments that using a noncomplementary DNA were also carried out. Our results showed that PNA can induce AuNP aggregation even with the presence of interfering ssDNAs (Fig. 3b, spectrum e). The fluorescence intensity of the mixture is restored to an extent that is comparable with PNA alone (Fig. 3a, spectrum e). The presence and absence of the target DNA can also be discriminated through solution color changes as we reported previously [41] (Fig. 3c). Based on these results, it can be deduced that the PNA-regulated FRET biosensor can be used for specific DNA determination.
a Fluorescence spectra of (a) CDs, (b) CD/AuNP, (c) CD/AuNP with PNA, (d) CD/AuNP with PNA-DNA complexes, (e) CD/AuNP with PNA and a random sequence DNA. b UV-vis absorption spectra and c photographs of (a) AuNPs, (b) CD/AuNP, (c) CD/AuNP with PNA, (d) CD/AuNP with PNA-DNA complexes, (e) CD/AuNP with PNA and a random sequence DNA
Optimization of assay conditions
To achieve better sensitivity and specificity of this method, the following parameters are optimized: (a) AuNP concentration; (b) PNA concentration; (c) NaCl concentration. Detailed results and discussions on optimizations are given in the Electronic Supporting Material (Fig. S5). In short, the following experimental conditions were found to give best results: (a) Best AuNP concentration: 5 nM; (b) Best PNA concentration: 0.8 μM; (c) Best NaCl concentration: 5 mM. It is worth noting that the stability of AuNPs is susceptible to the ionic strength, and the assay should be carried out in a low-salt buffer.
DNA analysis
Under the optimal experiment conditions, a piece of the human tumor suppressor gene TP53 DNA was used as the target to examine the applicability of this PNA-regulated FRET method. In order to quantify target DNA, the fluorescence quenching efficiency (F0 − F)/F0 is employed to evaluate the assay performance, where F0 represents the fluorescence intensity of CDs in the presence of AuNPs and PNA and F represents the fluorescence intensity of CDs in the presence of AuNPs and PNA after addition of DNA. As shown in Fig. 4a, the fluorescence intensity emitted by CDs decreases gradually with increasing DNA concentration. This decrease is attributed to the formation of PNA/DNA hybrids, which can stabilize AuNPs in solution and lead to further fluorescence quenching of CDs. A linear correlation exists between the fluorescence quenching efficiency and the concentration of DNA target over the range of 5–100 nM (R2 = 0.995) (Fig. 4b). This method has a detection limit of 0.21 nM based on 3σ/slope (where σ is the standard deviation). Compared with the other reported FRET-based methods for DNA detection, this PNA-regulated AuNP/CD FRET platform has a comparable sensitivity or a wider linear range (Table 1). Significantly, the probes and nanomaterials do not need to be labeled and modified.
a Fluorescence emission spectra of CDs in the presence of 5 nM AuNPs and 0.8 μM PNA with addition of different concentrations of target DNA (0, 5, 10, 20, 30, 40, 60, 80, 100, 150 nM). b The corresponding plot of (F0 − F)/F0 versus the concentration of target DNA. Error bars estimated as the standard deviation of three replicates
Specificity of the DNA assay
As demonstrated above, the PNA-regulated FRET assay is capable of discriminating between perfectly complementary DNA and random DNA (see Fig. 3). To further evaluate the performance of the method for specific DNA detection, three single-base mismatched targets with three different mismatch positions across the sequence (see Table S1) were designed and used for detection. As can be seen in Fig. 5 and Fig. S6, the fluorescence quenching efficiency is larger for the fully matched DNA than for the single-base mismatched ones, and significant differences are observed for Mutant 4 (p < 0.01) and Mutant 8 (p < 0.05). It is interesting that the DNAs, containing a mismatch located near the middle of the target, have the lower fluorescence quenching efficiencies, which makes it possible locate the mutation position of the target based on these observations. These results demonstrate that the FRET method is highly selective that can effectively distinguish a range of mismatched DNA targets from the fully matched ones.
Fluorescence quench efficiency changes with addition of 100 nM fully matched DNA or single-base mismatched DNAs (see Table S1). Error bars estimated as the standard deviation of three replicates. t test: ***p = 0.004, **p = 0.04, *p = 0.12
Determination of DNA in spiked samples
To demonstrate the practicality of the FRET method for biological samples, 10% of HeLa cell lysate samples spiked with different concentration of target DNA were examined. As displayed in Fig. 6, the fluorescence quenching efficiency increases with the increase in target concentration, and the fluorescence signals show a clear linear dependence on the target concentration from 10 to 100 nM. The calculated detection limit for the full-matched target in 10% cell lysate is 3.5 nM, which is comparable with the sensitivity in buffer. These results suggest the possibility of applying this strategy for specific DNA determination in biological samples.
To validate the target determination, recovery assays were performed by spiking known amount of target DNA into 10% cell lysate, and the results listed in Table S2 shows that the recovery is in the range of 96.5–102.4% with relative standard deviations (RSDs) less than 7.4%. This indicates the reliability of the method for the determination of DNA in biological samples.
Conclusion
We have developed a dual-mode DNA assay with both fluorescent and colorimetric readout based on PNA-regulated FRET between CDs and AuNPs. Free PNA is able to cause immediate aggregation of AuNPs, while PNA-DNA complexes can effectively stabilize unmodified AuNPs. FRET efficiency between CDs and AuNPs is affected by the aggregation/dispersion state of AuNPs. Therefore, after introducing of complementary DNA, the fluorescence quenching of CDs was enhanced due to the increased FRET between CDs and dispersed AuNPs. Based on the mechanism, the method allows rapid and sensitive determination of target DNA. This method is able to effectively discriminate a range of single-base mismatched DNAs from fully matched ones. Overall, this assay is convenient to operate without complicated modification or labeling steps of nanomaterials and probes, which make this method robust, cost-efficient, and thus promising for rapid DNA testing in clinical diagnosis. Theoretically, this FRET strategy not only allows the determination of nuclei acids but also provides the possibility for the analysis of other substances, which can directly or indirectly regulate the aggregation and dispersion of AuNPs. Future studies will focus on the further improvement of the sensitivity through signal amplification strategy and the performances of simultaneous determination of multiple analytes in real samples to exploit its full potential.
References
Bidard F-C, Weigelt B, Reis-Filho JS (2013) Going with the flow: from circulating tumor cells to DNA. Sci Transl Med 5:207–214
Fiala C, Diamandis EP (2018) Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection. BMC Med 16:166
Yuan L, Lin W, Zheng K, He L, Huang W (2013) Far-red to near infrared analyte-responsive fluorescent probes based on organic fluorophore platforms for fluorescence imaging. Chem Soc Rev 42:622–661
Yang Y, Zhao Q, Feng W, Li F (2013) Luminescent chemodosimeters for bioimaging. Chem Rev 113:192–270
Wang L, Dong L, Liu G, Shen X, Wang J, Zhu C, Ding M, Wen Y (2019) Fluorometric determination of HIV DNA using molybdenum disulfide nanosheets and exonuclease III-assisted amplification. Mikrochim Acta 186:286
Mandal TK, Parvin N, Mishra K, Mohandoss S, Lee YR (2019) Sensitive and selective fluorometric determination of DNA by using layered hexagonal nanosheets of a covalent organic framework prepared from p-phenylenediamine and benzene-1,3,5-tricarboxaldehyde. Microchim Acta 186:833
Iwe I, Li Z, Huang J (2019) Graphene oxide and enzyme-assisted dual-cycling amplification method for sensitive fluorometric determination of DNA. Microchim Acta 186:716
Tyagi S, Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14:303–308
Yang CYJ, Medley CD, Tan WH (2005) Monitoring nucleic acids using molecular beacons. Curr Pharm Biotechnol 6:445–452
Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol 16:359–363
Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang L (2005) Real-time detection of gene expression in cancer cells using molecular beacon imaging: new strategies for cancer research. Cancer Res 65:1909–1917
Radi AE, Sanchez JLA, Baldrich E, O'Sullivan CK (2006) Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J Am Chem Soc 128:117–124
Boens N, Leen V, Dehaen W (2012) Fluorescent indicators based on BODIPY. Chem Soc Rev 41:1130–1172
Lim SY, Shen W, Gao Z (2015) Carbon quantum dots and their applications. Chem Soc Rev 44:362–381
Sun Y-P, Zhou B, Lin Y, Wang W, Fernando KAS, Pathak P, Meziani MJ, Harruff BA, Wang X, Wang H, Luo PG, Yang H, Kose ME, Chen B, Veca LM, Xie S-Y (2006) Quantum-sized carbon dots for bright and colorful photoluminescence. J Am Chem Soc 128:7756–7757
Baker SN, Baker GA (2010) Luminescent carbon nanodots: emergent nanolights. Angew Chem Int Ed 49:6726–6744
Xu Q, Gong Y, Zhang Z, Miao Y, Li D, Yan G (2019) Preparation of graphene oxide quantum dots from waste toner, and their application to a fluorometric DNA hybridization assay. Microchim Acta 186:483
Tang J, Kong B, Wu H, Xu M, Wang Y, Wang Y, Zhao D, Zheng G (2013) Carbon nanodots featuring efficient FRET for real-time monitoring of drug delivery and two-photon imaging. Adv Mater 25:6569–6574
Loo AH, Sofer Z, Bousa D, Ulbrich P, Bonanni A, Pumera M (2016) Carboxylic carbon quantum dots as a fluorescent sensing platform for DNA detection. ACS Appl Mater Interfaces 8:1951–1957
Li F, Pei H, Wang L, Lu J, Gao J, Jiang B, Zhao X, Fan C (2013) Nanomaterial-based fluorescent DNA analysis: a comparative study of the quenching effects of graphene oxide, carbon nanotubes, and gold nanoparticles. Adv Funct Mater 23:4140–4148
Shi J, Tian F, Lyu J, Yang M (2015) Nanoparticle based fluorescence resonance energy transfer (FRET) for biosensing applications. J Mater Chem B 3:6989–7005
Lee MH, Leu CC, Lin CC, Tseng YF, Lin HY, Yang CN (2019) Gold-decorated magnetic nanoparticles modified with hairpin-shaped DNA for fluorometric discrimination of single-base mismatch DNA. Microchim Acta 186:80
Bu D, Zhuang H, Yang G, Ping X (2014) An immunosensor designed for polybrominated biphenyl detection based on fluorescence resonance energy transfer (FRET) between carbon dots and gold nanoparticles. Sensors Actuators B Chem 195:540–548
Shi Y, Pan Y, Zhang H, Zhang Z, Li M-J, Yi C, Yang M (2014) A dual-mode nanosensor based on carbon quantum dots and gold nanoparticles for discriminative detection of glutathione in human plasma. Biosens Bioelectron 56:39–45
Deng J, Lu Q, Hou Y, Liu M, Li H, Zhang Y, Yao S (2015) Nanosensor composed of nitrogen-doped carbon dots and gold nanoparticles for highly selective detection of cysteine with multiple signals. Anal Chem 87:2195–2203
Wang B, Chen Y, Wu Y, Weng B, Liu Y, Lu Z, Li CM, Yu C (2016) Aptamer induced assembly of fluorescent nitrogen-doped carbon dots on gold nanoparticles for sensitive detection of AFB1. Biosens Bioelectron 78:23–30
Qu F, Huang W, You J (2018) A fluorescent sensor for detecting dopamine and tyrosinase activity by dual-emission carbon dots and gold nanoparticles. Colloids Surf B: Biointerfaces 162:212–219
Li J, Rao X, Xiang F, Wei J, Yuan M, Liu Z (2018) A photoluminescence "switch-on" nanosensor composed of nitrogen and sulphur co-doped carbon dots and gold nanoparticles for discriminative detection of glutathione. Analyst 143:2083–2089
Yang Y, Huo D, Wu H, Wang X, Yang J, Bian M, Ma Y, Hou C (2018) N, P-doped carbon quantum dots as a fluorescent sensing platform for carbendazim detection based on fluorescence resonance energy transfer. Sensors Actuators B Chem 274:296–303
Yang Y, Hou J, Huo D, Wang X, Li J, Xu G, Bian M, He Q, Hou C, Yang M (2019) Green emitting carbon dots for sensitive fluorometric determination of cartap based on its aggregation effect on gold nanoparticles. Microchim Acta 186:259
Qin X, Lu Y, Bian M, Xiao Z, Zhang Y, Yuan Y (2019) Influence of gold nanoparticles in different aggregation states on the fluorescence of carbon dots and its application. Anal Chim Acta 1091:119–126
Xu S, Zhang F, Xu L, Liu X, Ma P, Sun Y, Wang X, Song D (2018) A fluorescence resonance energy transfer biosensor based on carbon dots and gold nanoparticles for the detection of trypsin. Sensors Actuators B Chem 273:1015–1021
Wang W, Wang Y, Pan H, Cheddah S, Yan C (2019) Aptamer-based fluorometric determination for mucin 1 using gold nanoparticles and carbon dots. Microchim Acta 186:544
Zhong D, Yang K, Wang Y, Yang X (2017) Dual-channel sensing strategy based on gold nanoparticles cooperating with carbon dots and hairpin structure for assaying RNA and DNA. Talanta 175:217–223
Qaddare SH, Salimi A (2017) Amplified fluorescent sensing of DNA using luminescent carbon dots and AuNPs/GO as a sensing platform: a novel coupling of FRET and DNA hybridization for homogeneous HIV-1 gene detection at femtomolar level. Biosens Bioelectron 89:773–780
Krawczak M (1990) The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet 85:55–74
Krawczak M, Cooper DN (1996) Single base-pair substitutions in pathology and evolution: two sides to the same coin. Hum Mutat 8:23–31
Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NST, Abeysinghe S, Krawczak M, Cooper DN (2003) Human gene mutation database (HGMD (R)): 2003 update. Hum Mutat 21:577–581
Egholm M, Buchardt O, Nielsen PE, Berg RH (1992) Peptide nucleic-acids (PNA)-oligonucleotide analogs with an achiral peptide backbone. J Am Chem Soc 114:1895–1897
Su X, Kanjanawarut R (2009) Control of metal nanoparticles aggregation and dispersion by PNA and PNA-DNA complexes, and its application for colorimetric DNA detection. ACS Nano 3:2751–2759
Xing S, Xu X, Fu P, Xu M, Gao T, Zhang X, Zhao C (2019) Colorimetric detection of single base-pair mismatches based on the interactions of PNA and PNA/DNA complexes with unmodified gold nanoparticles. Colloids Surf B: Biointerfaces 181:333–340
Xu W, Xing S, Xu X, Xu M, Fu P, Gao T, Zhao C (2018) Peptide nucleic acid-assisted label-free detection of single-nucleotide polymorphisms based on light scattering of carbon nanotubes. ACS Omega 3:17835–17841
Zhao C, Hoppe T, Setty MKHG, Murray D, Chun T-W, Hewlett I, Appella DH (2014) Quantification of plasma HIV RNA using chemically engineered peptide nucleic acids. Nat Commun 5:5079
Grabar KC, Freeman RG, Hommer MB, Natan MJ (1995) Preparation and characterization of au colloid monolayers. Anal Chem 67:735–743
Zeng P, Hou P, Jing CJ, Huang CZ (2018) Highly sensitive detection of hepatitis C virus DNA by using a one-donor-four-acceptors FRET probe. Talanta 185:118–122
Bao B, Pan Y, Gu B, Chen J, Xu Y, Su P, Liu Y, Tong L, Wang L (2018) Highly sensitive detection of nucleic acids using a cascade amplification strategy based on exonuclease III-assisted target recycling and conjugated polyelectrolytes. Analyst 143:4267–4272
Meng Y, Liu P, Zhou W, Ding J, Liu J (2018) Bioorthogonal DNA adsorption on polydopamine nanoparticles mediated by metal coordination for highly robust sensing in serum and living cells. ACS Nano 12:9070–9080
Li Y, Sun L, Qian J, Long L, Li H, Liu Q, Cai J, Wang K (2017) Fluorescent "on-off-on" switching sensor based on CdTe quantum dots coupled with multiwalled carbon nanotubes@graphene oxide nanoribbons for simultaneous monitoring of dual foreign DNAs in transgenic soybean. Biosens Bioelectron 92:26–32
Qian ZS, Shan XY, Chai LJ, Ma JJ, Chen JR, Feng H (2014) DNA nanosensor based on biocompatible graphene quantum dots and carbon nanotubes. Biosens Bioelectron 60:64–70
Funding
This work was supported by Ningbo Natural Science Foundation (2017C110020, 2018A610318, 2019C50039) and funds from Ningbo Institute of Materials Technology and Engineering, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 16686 kb)
Rights and permissions
About this article
Cite this article
Gao, T., Xing, S., Xu, M. et al. A peptide nucleic acid–regulated fluorescence resonance energy transfer DNA assay based on the use of carbon dots and gold nanoparticles. Microchim Acta 187, 375 (2020). https://doi.org/10.1007/s00604-020-04357-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04357-w