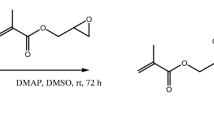
Abstract
An approach is presented for covalent immobilization of biomolecules on an acrylate phosphorylcholine hydrogel. The immobilization and the hydrogel formation take place simultaneously by a thiol-acrylate coupling reaction, induced by UV-light (254 nm). The hydrogel is prepared on two polymeric surfaces (the HardCoat protective layer of Blu-Ray discs, and SU-8) and applied to fluorescence microarray and label-free interferometric detection. For the first, Cy5 labeled analytes are used (λem 635 nm) and, for the second, a periodic array of high-aspect ratio nanopillars detects unlabeled analytes by interferometry. Bioavailability of the immobilized probes is demonstrated in labeled assays; for the case of oligonucleotides by discriminating single nucleotide polymorphisms, and, for the case of antibodies, by BSA immunorecognition. The raw hydrogel is employed to detect human C-reactive protein, in both labeled and non-labeled assay formats, with sensitivities of 30 ng·mL−1 and 2 pg·mL−1, respectively.

Schematic presentation of the phosphorylcholine (MPC) hydrogel preparation onto BluRay disc and SU-8 nanopillars to perform fluorescence and label-free interferometric detection, respectively. It selectively detects C-reactive protein (CRP), but it can covalently immobilize antibodies or nucleid acid probes to detect other analytes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Microarrays have found application in fields as health, food, environmental monitoring, and drug discovery among others [1]. It is mainly due to its multiplex and high-throughput screening capabilities. Regardless the progress done in microarrays during the last years, there are still challenges to overcome such as to improve sensitivity, selectivity and reproducibility, or to reduce the time of analysis, also. At this point, the immobilization of probes (proteins or nucleic acids) is a key process which needs to be re-designed for each support surface.
An appealing option is to use a hydrogel as a support for the immobilization of probes [2]. Hydrogels are cross-linked networks that retain a large amount of liquid in their swollen state but still maintain their 3D structure. They combine a soft nature and a porous structure that makes them ideal to immobilize proteins, enzymes or peptides [3]. For that, they have been used to mimic the physiologic environment and to increase the loading capacity [4]. Hydrogels have the potential to provide high signal-to-background ratios, good probe loading capability and low nonspecific binding [5, 6].
Having procedures providing simultaneously the hydrogel formation and the covalent attachment of the bioreceptor are undoubtedly desirable. But, very few examples are found in the literature [7]. A novel surface-bound in situ hydrogel for biosensing was developed by Herrmann et al. [8]. In there, the biomolecule is already present while the gel is formed from two different building blocks. This hydrogel is built up very fast and permits to covalently immobilize small molecules by the alkyne azide cycloaddition reaction. Nevertheless, the need on synthesizing all the building blocks, and modifying the probes makes this approach cumbersome. Thus, developing easy approaches, where the biomolecules are covalently attached to the hydrogel while it is formed, is still of interest.
Other important aspect is to avoid nonspecific adsorptions, in this sense hydrogels containing phosphorylcholine units can improve considerably the performance of the support. Phosphocholine is the polar, hydrophilic head group of some phospholipids found in cell’s plasma membrane. Besides, phosphocholine is the natural receptor of C-reactive protein, a pentraxin protein used as marker in inflammatory processes associated to cardiovascular diseases (CVD) [9]. Due to its biocompatibility, hygroscopicity and anti-fouling properties, methacrylate derivative of phosphorylcholine has been used to produce polymers with application as bulk materials for contact lenses, as coatings for medical devices such as coronary stents and, as additives in cosmetics.
Many methods are known for determination of C-reactive protein (CRP), most of them based on turbidimetry and/or immunoassay [10]. Interesting approaches such as immunoelectrochemical impedance spectroscopy [11] or immunochromatographic reactive strips [12] are highly competitive, but all them use antibodies as a capture probes. Very few examples are found using phosphocholine as a bioreceptor, although it presents the advantages of reducing 10-fold the reagent costs and to be more resistant and stable [13, 14]. Most of the examples uses different derivatives of phosphocholine to modify gold surfaces and to perform plasmon-based detection, reaching good sensitivities [15,16,17]. And, as far as we know, the use of phosphorylcholine hydrogel for microarray-based detection is not demonstrated yet, and neither on other label-free optical detection approaches, such as interferometry.
Here we propose the use of an acrylate derivative of phosphocholine which forms a hydrogel in aqueous solution simply under UV light irradiation, and, at the same time it can covalently attach thiolated probes (antibodies and nucleic acids) by means of a thiol-acrylate coupling chemistry. The method is easy, clean, and rapid, and proceeds in aqueous media. The irradiation also promotes the hydrogel attachment to the solid support. The employed materials are of polymeric nature, it includes Blu-ray discs, modified on their protective layer, for microarray fluorometric detection; and SU-8 surfaces, for label-free optical detection. Blu-ray discs were selected as microarray supports for their appealing properties such as stability, low unspecific adsorption, low background signal, compatibility with cheap detection systems, low cost, availability at mass scale, and good resistance to heat and impact [18]. SU-8 was the material constituting the optical transducer named Biophotonic Sensing Cell, BICELL, which is based in periodic nanopillars as described in previous works [19,20,21]. In this transducer the interferometric peak in the IR range (9000 to 16000 cm−1) decreases in wavelength as the biomolecule concentration increases on the surface. It presents the advantages of high limits of detection, vertical light coupling and multiplex capability. BICELLs constitutes the transducers here used to demonstrate the potential of our hydrogel to be used in sensitive label-free biosensing.
In this work, we demonstrate the different uses of the hydrogel. Firstly we apply it to fabricate a microarray for fluorometric CRP detection. In this case, the hydrogel will provide a friendly environment for the CRP binding. And also, it will increase the number of receptors in the spot. At the same time, the phosphorylcholine will block non-specific adsorption of other proteins present in the matrix (typically blood serum), due to its repellant property. Secondly, we employ the hydrogel as a network to create microarrays were oligonucleotide probes or antibodies are immobilized and the biointeractions take place successfully inside the hydrogel. Finally, we use the hydrogel to perform interferometric label-free detection of CRP in blood serum using BICELLs.
Materials and methods
Chemicals
Albumin from bovine serum ≥98% lyophilized powder (BSA), anti-Bovine Albumin antibody produced in rabbit (anti-BSA), C reactive protein from human plasma solution (in 0.02 M Tris, 0.28 M sodium chloride, 0.005 M calcium chloride, pH 7.8–8.2, containing 0.1% sodium azide), lactate dehydrogenase antibody (a-LDH), goat anti-rabbit immunoglobulin (GAR), Alexa-labeled goat anti-rabbit immunoglobulin (GAR-Alexa), Trizma base, and 2-methacryloyloxyethyl phosphorylcholine were purchased from Sigma-Aldrich Quimica (Spain, www.sigmaaldrich.com/spain.html). The oligonucleotides used in different experiments are listed in Table 1 and were provided by Sigma-Genosys (UK, www.sigmaaldrich.com/life-science/custom-oligos.html). Alexa Fluor® 647 NHS Ester (Succinimidyl Ester) was supplied by ThermoFisher (Spain, www.thermofisher.com). Buffers employed: phosphate buffered saline (PBS1x, 0.008 M disodium phosphate, 0.002 M monosodium phosphate, 0.137 M sodium chloride, 2.7 M potassium chloride, pH 7.5), PBS-T (PBS1x containing 0.05% Tween 20), carbonate buffer (CB, 0.015 M sodium carbonate, 0.035 M sodium bicarbonate, pH 9.6), SSC10x (1.5 M sodium chloride and 0.15 M sodium citrate, pH 7) Tris Buffer (1 M, pH 4). All the buffers and washing solutions were filtered through a 0.22 μm pore size nitrocellulose membrane from GE Whatman (www.sigmaaldrich.com) before use. Purification of anti-BSA antibody was done with an affinity Column Hi Trap Protein-A HP from GE Healthcare (Spain, www.gehealthcare.com), BSA, anti-BSA and CRP were labeled with Alexa647 in the lab following the supplier’s instructions. The name and sequence of the employed oligonucleotides can be found in Table S1, ESI.
Instrumental techniques
Microarray printing was carried out with a low volume, non-contact robotic dispensing system Biodot AD1500 (USA, www.biodot.com). Water contact angle measurements were carried out with Attension Theta Lite and the images treated with OneAttension (version 3.1) both from Biolin (Spain, www.biolinscientific.com). The measurements were done in triplicate at room temperature with a volume drop of 5 μL employing 18 mΩ water quality. UV-Ozone Cleaning System UVOH150 LAB from FHR (Germany, www.fhr.biz), equipped with a UV grid lamp (254 nm, 50 mW·cm−2) was employed for surface activation and for hydrogel formation and probe attachment. The protein solutions were concentrated and purified in a Centrifuge 5412D from Eppendorf (Spain, www.eppendorf.com) with Pierce Protein Concentrator PES (10 K, 30 K and 100 K MWCO) from ThermoFisher (Spain, www.thermofisher.com). Blu-ray Disk MediaRange were provided by the CD Rohling-up house GmbH (Germany, www.rohling-express.com). Concentrations of proteins were determined by measuring the optical density at 280 nm with a NanoDrop 2000 spectrophotometer from Thermo Scientific (Spain, www.thermofisher.com). The fluorescence signal of the spots was measured using a homemade surface fluorescence reader (SFR). [22]. Measurements were made by accumulation of emitted light (633 nm) by the samples during 8 s with a device gain of 3. For standard microarray image analysis, GenePix Pro 4.0 software from Molecular Devices, Inc. (USA, www.moleculardevices.com) was employed. Fluorescence Confocal Microscopy measurements were done with a Leica TCS-SP2-AOBS microscope at 633 nm excitation and 670 nm emission laser lines, with a diode detector for interferential contrast, a motorized platine and an incubation chamber to regulate the temperature and CO2. X-ray photoelectron spectroscopy was performed with SEGE 150 equipment from SPECS GMBH-Surface Analysis (Germany, www.specs-group.com).
Experimental procedures
Solid support preparation
Two different solid supports were employed in this paper. Blu-Ray disc surfaces, to develop hydrogel based fluorescence microarrays, and SU-8 surfaces to develop interferometric label-free detection. As an intermediate step, the optimization of the hydrogel formation onto SU-8 material was done employing glass slides with SU-8 on top and using fluorescence microarray format.
For Blu-Ray disc (BD) microarrays, BD were used as originally after washing with isopropanol and deionized water and drying. When necessary they were cut into slides of 5 × 2.5 cm.
For SU-8 microarrays, microscope glass slides (Labbox, Barcelona, Spain) were treated with piranha solution (H2O2:H2SO4 1:3 v/v), rinsed with deionized water and ethanol, and blow dried. The chips were pre-heated at 80 °C for 1 min and then 1 mL of SU-8 for each 5 cm2 of substrate was deposited in the center of slide. After spinning at 4000 rpm for 50 s, the slides were baked at 80 °C for 1 min and irradiated with UV light in flood exposure from mask aligner MA6 from Süss ensuring a full exposure of the resist. Then, they were baked at 80 °C for 1 min for full crosslinking and immersed in developer solution for 2 min in order to assure that no crosslinked resist remains. Finally, they were cut into smaller pieces (1 × 1 cm) using a tungsten awl.
Hydrogel microarray formation and determination of C-reactive protein (CRP)
2-Methacryloyloxyethyl phosphorylcholine (MPC) was purified by chromatography (Inhibitor remover, Sigma-Aldrich) to eliminate the MEQH inhibitor. Then, 14.75 g were dissolved in 10 mL of deionized water to obtain a 5 M solution. Dilutions from 0.5 to 3 M were done and arrays of 5 × 4 (30 nL/spot) were spotted onto solid support using a non-contact printing robotic arrayer (Biodot, Irvine, CA, USA). Chips were then irradiated for 7 min at 254 nm, incubated in dark for 10 min, water washed and air dried.
C-reactive protein from human plasma was labeled with Alexa647 dye following the protocol recommended by the supplier. 200 μL of CRP at 166 μg·mL−1 in PBS were obtained with a labeling ratio (Alexa/CRP) of 2. After, aliquots of CRP in PBS-T or in blood serum from rabbit (from 0 to 10 μg·mL−1) were incubated over the microarrays for 30 min at room temperature in humid chamber. Finally, the microarrays were washed by immersion for 15 min in PBS-T and 5 min in water, and air dried.
Fluorescence was quantified by the surface fluorescence reader (SFR) and images treated with GenePix software. Due to the illumination of the SFR, the accuracy in the measurement depends on the position of the row in the microarray, and thus error bars result dependent also on this fact.
Antibody immobilization in the MPC hydrogel and protein detection
Several solutions of anti-BSA were prepared at different concentrations from 0 μg·mL−1 to 100 μg·mL−1 in MPC 1 M (PBS1x). They were spotted onto the BD chips (4 spots/row, 30 nL) and irradiated at 254 nm for 40 s. The surface was water washed and air dried.
BSA was labeled with Alexa647 dye as before (labeling ratio = 2). Then, concentrations of labeled BSA ranging from 0 to 10 μg·mL−1 in PBS-T were incubated onto the microarrays for 30 min at room temperature. The chips were washed with PBS-t for 14 min, then with water and finally blow dried.
Fluorescence was measured and quantified as before.
Oligonucleotide probe immobilization in the MPC hydrogel and hybridization assays
Concentrations of P1 were prepared in MPC 1 M in water (from 0.1 to 5 μM). They were spotted onto the BD chips (4 spots/row, 30 nL) and irradiated at 254 nm for 7 min. The chips were washed for 15 min in PBS-T and for 10 min in water. The microarrays were incubated with decreasing concentrations of T1* in SSC5x (from 0.005 to 0.03 μM) for 1 h at 37 °C. Then, the chips were washed for 15 min in SSC1x and blow dried.
For the single nucleotide polymorphisms assay the immobilization conditions were the same and the hybridization was carried out in SSC1x with different percentages of formamide (0 to 50%).
BICELL fabrication
BICELLs were produced by contact lithography at wafer level using a mask aligner MA6. It was used 6” Si wafers as a substrate. A single lithography step was applied to produce the photonic structures.
First, ultrasonication baths on acetone and water were performed for 2 min. After rinsing and drying, treatment for 30 s in hydrofluoric acid diluted with water (HF:H2O, 1:10 v/v) was done, followed by ultrasonication in water for 2 min. Caution: HF is a hazardous product that should be handle with extreme care, in fume hood and wearing protective clothing. After rinsing and drying, Si wafer was immersed in piranha solution (H2SO4:H2O2, 2:1 v/v) for 20 min. Caution: piranha solutions react violently with organic materials and should be handled with extreme care. Finally, wafer was rinsed, dried and heated up to 120 °C for 1 h.
Then, SU-8 was deposited on the surface using a dilution of SU-8 2010 in cyclopentanone (CP). By controlling the spin-speed, SU-8 thicknesses of: 200 nm and ~ 0.9–1.2 μm were obtained.
Later a soft-bake for 30 s at 70 °C, exposure during 10 s (power density ~ 40 mW·cm−2) with no UV pass-band filter, by using photomasks of metal on quartz for contact lithography, post-bake during 60 s at 70 °C, development during 120 s, and rinse with isopropanol (IPA) and blown with dry compressed air were carried out. Finally, a hard-bake was performed during 10 min at 70 °C to harden the polymer.
Two types of BICELLs were produced: Fabry-Perot interferometers, based on a single thin-film, and periodic lattices of microstructures. Those photonic structures were concerned onto cells 200 × 200 μm in size.
Hydrogel formation and CRP recognition in the Fabry-Perot interferometer
10 μL of PC 0.5 M were deposited on the interferometer surface and irradiated at 254 nm for 60 s. After washing with water and drying with air, the optical response was measured.
To construct the calibration plot, chips were serially incubated with increasing concentrations of CRP in PBS-T (from 10 to 10000 ng·mL−1, 10 μL) for 30 min at 37 °C in humidity. After each incubation step the chip was washed extensively with PBS-T and water, and dried, and the optical response was measured by Fourier Transform Infrared-Visible (FTIR-VIS) spectrometer attached to a microscope for microdomain measurement whole 200 × 200 μm BICELLs.
For real samples measurements, blood serum was doped with 2 μg·mL−1 of CRP and incubated and measured as before.
Hydrogel formation and CRP response in the BICELLs
10 μL of PC 0.1 M were spread out over the BICELLs and irradiated at 254 nm. After washing with water and drying, the optical response was characterized similarly to Fabry-Perot interferometers.
Incubations with increasing concentrations of C-reactive protein in PBS-T were done as before to obtain the calibration plot. Human blood serum samples, with basal values of CRP, were collected from healthy volunteers. Blood sera were doped with 50 ng·mL−1 of CRP to determine the recovery percentage.
Discussion and results
Choice of materials
In this work a fluorescence microarray is set-up, using an acrylate hydrogel of phosphorylcholine for specific recognition of C-reactive protein. A major challenge of the read-out process is to obtain high fluorescence signal-to-background ratio in the areas where protein is captured. In addition, non-specific adsorption of other fluorescing molecules must be reduced to a minimum. Consequently, the choice of the materials for the chip is important.
BluRay disc (BD) surface was chosen for three reasons. Firstly, it is available at mass scale at a very low cost. Secondly, it has interesting physical-chemical properties such as stability, low unspecific adsorption, and resistance to scratching and heat. Thirdly, the protective layer of BD is vinyl or epoxy-based, which permits the covalent attachment of the acrylate monomer units to the surface, performing a robust link.
SU-8 material was selected because it is an epoxy-based resin, thus the hydrogels can be also covalently linked to the surface, and it is a common material to fabricate interferometric transducers by photolithography and other fabrication approaches. Here, the applicability of the hydrogel to biofunctionalized interferometric transducers made of SU-8 is demonstrated.
Hydrogel formation and C-reactive protein (CRP) detection in microarray format
CRP is a highly attractive biomarker for cardiac risk prediction, as well as a good indicator of inflammatory processes. Combining a derivative of o-phosphorylethylethanolamine with the use of a hydrogel must result in a detection system having enhanced performance in terms of sensitivity and easiness of use. Phosphorylethylethanolamine is specific towards CRP and does not cross react with albumin and cytochrome C. The hydrogel increases the number of receptors available compared to the “conventional” immobilization modes.
To optimize the microarray construction, purified 2-methacryloyloxyethylphosphorylcholine (MPC) was dissolved in different solvents (water, ethanol, tetrahydrofuran, and toluene) at 1 M concentrations. The solutions were then spotted on the chip surface and irradiated for different times (1 min to 1 h) at 254 nm. At this wavelength, the radical photopolymerization of the double bonds undergoes rapidly, and the radicals formed promote the tethering the polymeric hydrogel to the epoxy or alkenyl substrates. The arrays were washed extensively with water, and the consistency of the spots was visually inspected. The hydrogel performed in water under 7 min UV light irradiation provided the higher resistance and reproducibility, thus it was selected for further optimizations (Fig. 1).
The phosphorylcholine units provide a highly hydrophilic environment (WCA < 10o), in contrast with the BD (WCA 80°). This high hydrophilicity will favor the contact with the aqueous sample. At the same time, the well-known repellent properties of phosphocholine will hinder the unspecific adsorption of proteins in the spot, and only the CRP from the analyte sample will be bound through the specific interaction with PC.
Different concentrations of aqueous MPC, from 0.1 to 3 M, were microarrayed onto BD. The microarrays were used to characterize the response towards CRP recognition. For that a solution of Alexa647 labeled CRP was used. The first assays were done using PBS-T to dilute the CRP. Dilutions ranging from 0.075 μg·mL−1 to 5000 μg·mL−1 were spread over the microarray and incubated for 30 min at room temperature after washing the chips and processing the fluorescence, signal distinguishable of background was detected for all the assayed concentrations (ESI Fig. S1). As CRP recognition can be calcium dependent, new assays were done adding 10 mM Calcium Chloride to the binding buffer, PBS-T, without finding relevant improvement. A concentration of MPC 1 M was selected as the optimal for further assays.
As the final goal was to detect CRP in blood serum, new CRP solutions, from 0.08 to 10 μg·mL−1, diluted in blood serum instead of PBS-T were spread over new microarrays. The incubation time was 30 min at room temperature. All the concentrations were detected. The limit of detection, calculated as the concentration that provides the blank signal plus three times the standard deviation of blank, resulted 0.03 μg·mL−1. To demonstrate the specificity of the recognition, the same experiment was done, but using labeled bovine serum albumin (BSA) instead of CRP, no fluorescence signal was detected after washing the microarray. Also, a good linear relationship was observed between the fluorescence signal and the CRP concentration (Fig. 2, ESI Fig. S2 and S3). The sensitivity of our microarray was limited by the sensitivity of the homemade fluorescence reader, SFR. Considering the limitations of the reading device, the platforms revealed great suitability for the recognition of very low levels of protein, and, changing to other more sensitive detection device or using colorimetric or enzymatic detection can lead to limits of detection at least two orders of magnitude lower, as we previously demonstrated. [23]
C-reactive protein binding curve in blood serum for microarrays created onto BD using 1 M MPC. Inset: linear range and comparison to the assay performed using labeled BSA instead of CRP as a target. The hydrogel resulted specific to CRP. For the assays, AlexaFluor647 labeled CRP and BSA were used, and the fluorescence was measured with a homemade surface fluorescence reader (SFR) based on a CCD camera with LEDs as a excitation source (λexc 633 nm, λem 670 nm)
As the hydrogel has a 3D structure, fluorescence confocal microscopy was used to assess the penetration of the target in the spot. As can be seen in Fig. 3, the proteins penetrated inside the hydrogel by diffusion.
Image of a spot registered using Confocal Fluorescence microscopy for different z-planes and angles to assess the presence of fluorescence inside the spot. The labeled CRP penetrates inside the spot. On the right, image of the spot for transmitted light. For more detail see ESI video 1
To broaden the applicability of the hydrogel-based microarray, we studied the possibility of anchoring different probes with the hydrogel by means of the thiol-acrylate coupling click chemistry.
Antibody immobilization in the hydrogel and protein detection in microarray format
To analyze the capability of the hydrogel to immobilize antibodies and demonstrate their bioavailability, the model system BSA/anti-BSA (aBSA) was selected, as it is widely used, and extensive data are available in the literature for comparison.
Firstly, the photoimmobilization of the antibody was studied while hydrogel formation. It is hypothesized that the same irradiation that induces the acrylate polymerization also catalyzes the thiol-acrylate coupling reaction between MPC units and the antibody thiols. For that, antiBSA from rabbit serum was purified by a protein A affinity column and then it was labeled with Alexa647.
The following parameters were optimized: buffer, MPC concentration, washing conditions and aBSA concentration (from 1.56 μg·mL−1 to 100 μg·mL−1). Optimal conditions were found for PBS1x, 40 s of irradiation at 254 nm, and washings in PBS1x for 14 min after immobilization. Under these conditions, the fluorescence measured after washing was like that registered after irradiation. This indicated that all the aBSA spotted remained immobilized in the hydrogel (ESI, Fig. S4). The immobilization experiments were done in duplicate finding a RSD intrachip of 2–17% and a RSD interchip of 6–15%.
Considering the spotted volume and the spot diameter, immobilization densities ranged from 0.37 to 27.85 picomol·cm−2, for the lowest and the highest spotted concentrations, respectively. Under the saturation conditions (concentrations over 25 μg·mL−1), the immobilization density results approximately 10-fold the immobilization density expected for a close-packed monolayer of antibodies immobilized in a 2D surface (2.47–2.93 picomol·cm−2) [24].
To assess the antibody’s bioavailability immobilized inside the hydrogel, new microarrays were created as before but using non-labeled aBSA. The microarrays were subjected to incubation with increasing concentrations of Alexa647 labeled BSA in PBS-T (0 to 10 μg·mL−1) for 30 min at room temperature. The binding curves are shown in Fig. 4.
From the corresponding calibration plot (ESI, Fig. S5) it was possible to determine the amount of recognized BSA and relate it to the immobilized antibody. Biorecognized BSA densities ranged from 0.20 to 5.27 picomol·cm−2, which corresponds to biorecognition yields from 54% to 20%. In the microarray, a row of the MPC hydrogel without aBSA was incorporated to check that the BSA was retained inside the hydrogel because of the antibody’s specific recognition. As can be seen in Fig. 4, no fluorescence signal was detected in the case of the blank.
As for the case of CRP, fluorescence confocal microscopy analysis demonstrated the penetration of the BSA inside the spots (ESI Fig. S6 and video 2). The stability of the microarrays was assessed by preparing several chips and running the assay immediately and after 4, 7, 15 and 30 days storing the chips in the fridge. No significant loose of fluorescence was observed (ESI, Fig. S7).
Nucleic acid immobilization in the hydrogel and hybridizations in microarray format
Finally, we aimed to demonstrate the feasibility of the MPC hydrogel to immobilize nucleic acid probes and to run successful hybridizations in the inside of the hydrogel. For that commercially available thiol-modified oligonucleotides were cross-linked with polymer through thiol-acrylate coupling reaction, as in the case of the antibodies (see ESI Table S1).
Thus, solutions of 1 MPC in PBS1x containing different concentrations of fluorescence labeled oligonucleotides: P2* (thiol-ended), C2* (amine-ended) and T1* (phosphate-ended), were spotted on the BD and irradiated for 7 min at 254 nm. Then, fluorescence was measured after extensive washing. Only P2* remained inside the hydrogel demonstrating the need of the thiol moiety to achieve the covalent attachment by the thiol-acrylate coupling (ESI, Fig. S8).
To test the hybridization efficiency of the microarrays, newly ones were prepared including P3 (thiol-ended), P1 (thiol-ended), C2 (amine-ended) and P2* (thiol-ended). P1 and C2 had the sequence complementary to the target T1*, P3 was included to test the specificity of the hybridization and P2* as a probe immobilization control. After irradiation and washing, the chips were hybridized with T1* in SSC1x for 1 h at 37 °C. Then, they were thoroughly washed with SSC1x and measured with the SFR. As expected only the rows of the microarray corresponding to P1 and P2* provided significant fluorescence signal (Fig. 5).
Fluorescence signals for a microarray, designed as depicted in the figure inset, after hybridization with T1* in SSC1x for 1 h at 37 °C. T1* is complementary to sequences of P1, C2, and P2*. P2* was included as a positive immobilization control. Inset: fluorescence image of the microarray created spotting (4 spots/row, 30 nL/spot) MPC hydrogel, and MPC hydrogel having co-immobilized different probes (P1, P3, C2, P2*), and hybridized with labeled target T1*, only the rows having the probe complementary to T1* (P1) provided significant fluorescence signal
Different concentrations of T1* were assayed (from 0.5 to 0.003 μM), being able to detect up to 0.003 μM, which compares well with previous reported data (ESI, Fig. S9). LOD was determined as the concentration providing the blank signal plus three times the standard deviation of the blank. It resulted 6 nM, which correlates well with the lowest detected concentration for the microarrays. The saturation of hybridization is achieved with 1 μM of P1 and concentrations of T1* over 0.05 μM. The hybridization was also done in SSC 5x without improving the results for SSC1x. The fluorescence intensities are similar to that reported by our group previously using photo-click coupling chemistry without the aid of a hydrogel [25, 26], but in those cases the concentration of probe needed was between 5 and 10 μM while here this sensitivity is reached with probe concentrations of 0.1 and 0.5 μM. As said before, the sensitivity is limited by the performance of the SFR and can be lowered with a better detection device.
Finally, discrimination of single nucleotide polymorphism was attempted. For that, microarrays were prepared including probes P1, P1.1m, P1.2m, P1.3m and P3. They have the fully complementary sequence, one mismatch, two mismatches, three mismatches, and ten mismatches to T1*, respectively. The probes were immobilized at 5 μM, and the hybridization was run for 2 h at 37 °C, with T1* at 0.25 μM at different ionic strengths (SSC1x to SSC10x). For all the conditions successful discrimination was reached. When comparing to previous surfaces developed by our group, and performing the assays with the same sequences, it is seen that the MPC hydrogel permits a better discrimination between the perfect match and one mismatch sequence, even without astringency conditions (SSC1x) (Fig. 6, and ESI Fig. S10).
Relative fluorescence intensity signals for fully complementary (P1) and one mismatch (P1.1m) probes hybridizing with T1* (0.25 μM) in SSC1x for 1 h at 37 °C. Comparison between the performance reached for the same assay using hydrogel-based microarray (3D) and previously developed microarrays (2D) based on the thiol-yne [27], thiol-ene [28, 29] and thiol-fluor [26] coupling chemistry. The hydrogel-based microarray provides better SNP discrimination without employing astringency conditions
Label-free photonic sensing of C-reactive protein (CRP) by BICELLs
After that, we studied the capability of the hydrogel to be applied onto other surfaces or platforms containing epoxy or alkene groups. Thus, we tackled the biofunctionalization with MPC hydrogel of two types of interferometric structures made of SU-8 photoresist. The first transducer was a Fabry-Perot interferometer based on a 200 nm-thick layer of SU-8 onto Si. The second one was a Biophotonic sensing cell (BICELL) based on micro-nanopatterned structures (pillars) of SU-8 (Fig. 7b).
Sensing curves by label-free interferometric techniques wavelength shift (nm) vs CRP concentration (ng·mL−1) in PBS-T using Fabry-Perot (a). SEM image of the structure based on pillars, BICELLs (b). Sensing curves by label-free interferometric techniques wavelength shift (nm) vs CRP concentration (ng·mL−1) in PBS, comparison of Fabry-Perot and BICELLs structures (c)
Firstly, the biofunctionalization was done using fluorescence microarray format on SU-8-coated microscope glass slides. Then it was transferred to the interferometric microstructures to perform label-free biosensing. As occurred with BD supports, SU-8 showed very low fluorescence background and low unspecific adsorption. The formation of the MPC hydrogels onto SU-8 surfaces was optimized in a similar manner than it was done for the BD supports. As in the first case, hydrophilicity of the surface increased after hydrogel tethering, changing the WCA from 79o, for the raw SU-8, to 10o, for hydrogel biofunctionalized SU-8 (ESI Fig. S11). Also, the optimal concentration of MPC resulted 1 M, and biorecognition of fluorescence labeled CRP by the so created microarray provided a limit of detection of 0.025 μg·mL−1 (ESI Fig. S12 and S13).
After assessing the capability of the MPC hydrogels onto planar SU-8 substrates, the Fabry-Perot interferometer was functionalized with MPC using a modification of the protocol employed for the starting assay on microarray. Since the sensing event must be placed close to the surface, the amount of MPC was reduced to 10 μL at 0.5 M and irradiated under the same conditions than before. The optical response of the bare interferometer with the PC was registered by FTIR-VIS attached to a microscope in reflection mode with a 15X objective in the VIS-NIR spectral range. Afterwards, a wide range of concentrations of CRP in PBS-T, from ng·mL−1 to μg·mL−1 were prepared, and serially incubated on the Fabry-Perot structure. The optical response of the interferometer was measured after each incubation and washing step. The collected data were plotted to obtain the calibration plot (Fig. 7a).
Analyzing the responses between 0 and 25 ng·mL−1, the calculated sensitivity ms resulted 0.28 nm/ng·mL−1, which provided a LOD (Uw / ms) of 21 pg·mL−1. Here, the expanded uncertainty for the wavelength used for the measurements is Uw = 3 uw and the evaluation of wavenumber uncertainty (uw) during the optical interrogation process is uw2 = (wres)2 / 12 being wres the wavenumber resolution.
Finally, a new Fabry-Perot interferometer, functionalized with PC and optically characterized as before, was incubated with a sample of real blood serum, which was doped with 2 μg·mL−1, under the same conditions employed for the calibration assay. After washing, the optical response displayed a wavelength shift of 14 nm. This wavelength shift was interpolated in the fitted calibration plot, corresponding to a concentration of 1560 ng·mL−1 resulting on a recovery of 78%. Thus, undiluted blood serum may be used to detect CRP within significant clinical values.
BICELLs, consisting of periodic arrays of fractal structures based on different pillars with dimensions of d1 = 760 nm and d2 = 2 μm (Fig. 7b), were functionalized with MPC. This structure can improve the sensitivity with respect to a Fabry-Perot sensor due to the higher effective surface. The detailed principle of detection of BICELLs can be found elsewhere [19,20,21].
In this case, the concentration of PC employed was even lower than for the Fabry-Perot, and 0.1 M of MPC was spin coated onto the BICELLs and irradiated as before to create the hydrogel. Serial incubations with increasing concentrations of CRP in PBS-T were carried out as before, and the optical response was measured after each incubation and washing step. CRP concentrations ranged from 0 to 5000 ng·mL−1. To obtain the calibration plot the wavelength shift was plotted against the concentration of CRP. In Fig. 7c it can see the optical response for BICELL and Fabry-Perot structures in the same range of concentrations. The sensitivity reached for BICELL was 2.88 nm/ng mL−1, and therefore BICELLs structure improves in 10-fold the sensor sensitivity.
Undiluted blood serum was spiked with CRP at 50 ng·mL−1, and incubated on a BICELL, which was coated with PC as before. After characterizing its optical response, a wavelength shift of 45 nm was measured; corresponding to a concentration of approximately 40 ng·mL−1, interpolated in the corresponding calibration plot. In this case, the calculated recovery resulted 80%.
The CRP can be quantified with the interferometric structures in a wide range of concentrations without the need of modification or labeling. This approach reaches lower limit of detection than other reported label-free assays based most of them in SPR measurements. Thus, the LOD of 21 pg ·mL−1 reached with the Fabry-Perot structures overcomes the LOD reported for other impedimetric or plasmonic approaches, which are comparatively summarized in Table 1.
Thus, we here demonstrate that our hydrogel is a competitive manner to detect CRP which in combination with a proper transductor can reach high sensitivities. However, the most important point is that our hydrogel is a way to covalently immobilize a large number of bioreceptors (thiolated nucleic acids or antibodies) with good bioavailability. Besides, the hydrogel can be formed onto alkene or epoxy-ended surfaces, and can be applied to different detection formats (labeled or label-free).
Conclusion
In a simple and quick manner, a hydrogel is formed combining the photoinitiated polymerization of acrylate moieties with the presence of phosphorylcholine groups, which acts as antifouling agents, as well as specific receptors towards CRP. The procedure demonstrated to be an adequate method to biofunctionalize alkene or epoxy surfaces. MPC hydrogel proved excellent performance in immuno and oligonucleotide hybridizations assays. The immobilization method is fast, clean and biocompatible.
The hydrogel immobilization can be combined with different transductors working in labeled or label-free format. As an example, the combination of hydrogels with the BICELLs measurements principle proves to be a powerful tool for low abundance analytes.
The approach is especially interesting in complex matrix where unspecific adsorption can be a problem for the detection.
References
Rusmini F, Zhong Z, Feijen J (2007) Protein immobilization strategies for protein biochips. Biomacromolecules 86:1775–1789. https://doi.org/10.1021/BM061197B
Rubina AY, Dementieva EI, Stomakhin AA, Darii EL, Pan’kov SV, Barsky VE, Ivanov SM, Konovalova EV, Mirzabekov AD (2003) Hydrogel-based protein microchips: manufacturing, properties, and applications. Biotechniques 34:1008–1022. https://doi.org/10.2144/03345rr01
Vermonden T, Censi R, Hennink WE (2012) Hydrogels for Protein Delivery. Chem Rev 112:2853–2888. https://doi.org/10.1021/cr200157d
Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A (2000) Protein microchips: use for immunoassay and enzymatic reactions. Anal Biochem 278:123–131. https://doi.org/10.1006/ABIO.1999.4363
Moschallski M, Evers A, Brandstetter T, Rühe J (2013) Sensitivity of microarray based immunoassays using surface-attached hydrogels. Anal Chim Acta 781:72–79. https://doi.org/10.1016/j.aca.2013.04.013
Beyer A, Pollok S, Berg A, Weber K, Popp J (2014) Easy daylight fabricated hydrogel Array for colorimetric DNA analysis. Macromol Biosci 14:889–898. https://doi.org/10.1002/mabi.201300487
Kivlehan F, Paolucci M, Brennan D, Ragoussis I, Galvin P (2012) Three-dimensional hydrogel structures as optical sensor arrays, for the detection of specific DNA sequences. Anal Biochem 421:1–8. https://doi.org/10.1016/j.ab.2011.10.026
Herrmann A, Kaufmann L, Dey P, Haag R, Schedler U (2018) Bioorthogonal in situ hydrogels based on polyether polyols for new biosensor materials with high sensitivity. ACS Appl Mater Interfaces 10:11382–11390. https://doi.org/10.1021/acsami.8b01860
Thompson D, Pepys MB, Wood SP (1999) The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure 7:169–177. https://doi.org/10.1016/S0969-2126(99)80023-9
Algarra M, Gomes D, Esteves da Silva JCG (2013) Current analytical strategies for C-reactive protein quantification in blood. Clin Chim Acta 415:1–9. https://doi.org/10.1016/j.cca.2012.09.007
Boonkaew S, Chaiyo S, Jampasa S, Rengpipat S, Siangproh W, Chailapakul O (2019) An origami paper-based electrochemical immunoassay for the C-reactive protein using a screen-printed carbon electrode modified with graphene and gold nanoparticles. Microchim Acta 186:153. https://doi.org/10.1007/s00604-019-3245-8
Byzova NA, Zherdev AV, Vengerov YY, Starovoitova ТA, Dzantiev BB (2017) A triple immunochromatographic test for simultaneous determination of cardiac troponin I, fatty acid binding protein, and C-reactive protein biomarkers. Microchim Acta 184:463–471. https://doi.org/10.1007/s00604-016-2022-1
Kim E (2013) Protein-directed immobilization of Phosphocholine ligands on a gold surface for multivalent C-reactive protein binding. Curr Top Med Chem 13:519–524
Kitayama Y, Takeuchi T (2014) Localized surface Plasmon resonance Nanosensing of C-reactive protein with poly(2-methacryloyloxyethyl phosphorylcholine)-grafted gold nanoparticles prepared by surface-initiated atom transfer radical polymerization. Anal Chem 86:5587–5594. https://doi.org/10.1021/ac501322x
Kamon Y, Kitayama Y, Itakura AN, Fukazawa K, Ishihara K, Takeuchi T (2015) Synthesis of grafted phosphorylcholine polymer layers as specific recognition ligands for C-reactive protein focused on grafting density and thickness to achieve highly sensitive detection. Phys Chem Chem Phys 17:9951–9958. https://doi.org/10.1039/C5CP00469A
Yoshimoto J, Sangsuwan A, Osaka I, Yamashita K, Iwasaki Y, Inada M, Arakawa R, Kawasaki H (2015) Optical properties of 2-Methacryloyloxyethyl Phosphorylcholine-protected au 4 nanoclusters and their fluorescence sensing of C-reactive protein. J Phys Chem C 119:14319–14325. https://doi.org/10.1021/acs.jpcc.5b03934
Matsuura R, Tawa K, Kitayama Y, Takeuchi T (2016) A plasmonic chip-based bio/chemical hybrid sensing system for the highly sensitive detection of C-reactive protein. Chem Commun 52:3883–3886. https://doi.org/10.1039/C5CC07868G
Morais S, Tortajada-Genaro L, Maquieira A (2014) Array-on-a-disk? How Blu-ray technology can be applied to molecular diagnostics. Expert Rev Mol Diagn 14:773–775. https://doi.org/10.1586/14737159.2014.929945
Holgado M, Barrios C a., Ortega FJ, et al (2010) Label-free biosensing by means of periodic lattices of high aspect ratio SU-8 nano-pillars. Biosens Bioelectron 25:2553–2558. doi: https://doi.org/10.1016/j.bios.2010.04.042
Sanza FJ, Holgado M, Ortega FJ, Casquel R, López-Romero D, Bañuls MJ, Laguna MF, Barrios CA, Puchades R, Maquieira A (2011) Bio-photonic sensing cells over transparent substrates for anti-gestrinone antibodies biosensing. Biosens Bioelectron 26:4842–4847. https://doi.org/10.1016/j.bios.2011.06.010
Lavín Á, Casquel R, Sanza FJ, Laguna MF, Holgado M (2013) Efficient design and optimization of bio-photonic sensing cells (BICELLs) for label free biosensing. Sensors Actuators B Chem 176:753–760. https://doi.org/10.1016/j.snb.2012.09.058
Mira D, Llorente R, Morais S, et al (2004) <title>High-throughput screening of surface-enhanced fluorescence on industrial standard digital recording media</title> In: Carrano JC, Zukauskas A (eds). International Society for Optics and Photonics, pp 364–373
Alonso R, Jiménez-Meneses P, García-Rupérez J, Bañuls MJ, Maquieira Á (2018) Thiol–ene click chemistry towards easy microarraying of half-antibodies. Chem Commun 54:6144–6147. https://doi.org/10.1039/C8CC01369A
Chen H, Huang J, Lee J, Hwang S, Koh K (2010) Surface plasmon resonance spectroscopic characterization of antibody orientation and activity on the calixarene monolayer. Sensors Actuators B Chem 147:548–553. https://doi.org/10.1016/J.SNB.2010.03.033
Bañuls M-J, Jiménez-Meneses P, Meyer A, Vasseur JJ, Morvan F, Escorihuela J, Puchades R, Maquieira Á (2017) Improved performance of DNA microarray multiplex hybridization using probes anchored at several points by thiol–Ene or thiol–Yne coupling chemistry. Bioconjug Chem 28:496–506. https://doi.org/10.1021/acs.bioconjchem.6b00624
Jiménez-Meneses P, Bañuls M-J, Puchades R, Maquieira Á (2018) Fluor-thiol Photocoupling reaction for developing high performance nucleic acid (NA) microarrays. Anal Chem 90:11224–11231. https://doi.org/10.1021/acs.analchem.8b00265
Escorihuela J, Bañuls M-J, Puchades R, Maquieira Á (2014) Site-specific immobilization of DNA on silicon surfaces by using the thiol–yne reaction. J Mater Chem B 2:8510–8517. https://doi.org/10.1039/C4TB01108B
Escorihuela J, Bañuls MJ, Grijalvo S, Eritja R, Puchades R, Maquieira Á (2014) Direct covalent attachment of DNA microarrays by rapid thiol-ene “click” chemistry. Bioconjug Chem 25:618–627
González-Lucas D, Bañuls M-J, Puchades R, Maquieira Á (2016) Versatile and easy fabrication of advanced surfaces for high performance DNA microarrays. Adv Mater Interfaces 3:1500850. https://doi.org/10.1002/admi.201500850
Lee JH, Yoon KH, Hwang KS, Park J, Ahn S, Kim TS (2004) Label free novel electrical detection using micromachined PZT monolithic thin film cantilever for the detection of C-reactive protein. Biosens Bioelectron 20:269–275. https://doi.org/10.1016/j.bios.2004.01.024
Roberts WL, Moulton L, Law TC, Farrow G, Cooper-Anderson M, Savory J, Rifai N (2001) Evaluation of nine automated high-sensitivity C-reactive protein methods: implications for clinical and epidemiological applications. Part 2. Clin Chem 47:418–425
Hu WP, Hsu H-Y, Chiou A, Tseng KY, Lin HY, Chang GL, Chen SJ (2006) Immunodetection of pentamer and modified C-reactive protein using surface plasmon resonance biosensing. Biosens Bioelectron 21:1631–1637. https://doi.org/10.1016/j.bios.2005.11.001
Kim H-C, Lee S-K, Sohn Y-S, Ryu HK, Jeong SW (2007) Preparation of a sensing membrane for C-reactive protein. Macromol Symp 249–250:71–75. https://doi.org/10.1002/masy.200750312
Yuk JS, Jung J-W, Hyun J, Kim YM, Ha KS (2008) Development of a scanning surface plasmon microscope based on white light for analysis of a wide range of protein arrays. Sensors Actuators B Chem 131:241–246. https://doi.org/10.1016/j.snb.2007.11.019
Vashist SK, Schneider EM, Luong JHT (2015) Surface plasmon resonance-based immunoassay for human C-reactive protein. Analyst 140:4445–4452. https://doi.org/10.1039/C5AN00690B
Acknowledgements
This research was funded by FEDER and the MINECO projects CTQ/2016/75749-R and TEC/2017/84846-R. The authors acknowledge the contribution of the Bc’s D students Alejandra Vargas and Ismael Guerrero for the time spent at our lab and for their enthusiastic ideas on the possibilities of MPC hydrogel.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Díaz-Betancor, Z., Bañuls, MJ., Sanza, F.J. et al. Phosphorylcholine-based hydrogel for immobilization of biomolecules. Application to fluorometric microarrays for use in hybridization assays and immunoassays, and nanophotonic biosensing. Microchim Acta 186, 570 (2019). https://doi.org/10.1007/s00604-019-3691-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3691-3