Abstract
A specific and sensitive colorimetric aptasensor is described for the determination of Malachite Green (MG). It is exploiting the inhibition of the peroxidase-like activity of gold nanoparticles (AuNPs). The AuNPs act as enzyme mimics that catalyze the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) by H2O2 to yield a dark blue solution. The catalytic activity is inhibited by hexadecyl trimethyl ammonium ion, specifically by cetyltrimethylammonium bromide (CTAB), which causes the aggregation of AuNPs. If a (negatively charged) RNA-aptamer against MG is added, it binds to the positively charged CTAB and prevents aggregation. This enhances the enzyme mimicking activity of the AuNPs and leads to the formation of a dark blue solution. However, in the presence of MG, the aptamer binds to MG, and leads to the aggregation of AuNPs again. The aggregated AuNPs possess a light blue color. A colorimetric method (best performed at 650 nm) was work out that can detect MG in a concentration range from 10 to 500 nmol L−1. The detection limit based on 3σ/k criterion is 1.8 nmol L−1. The assay is highly specific and accurate. Recoveries from spiked real samples (aquaculture water) ranged from 80% to 120%.

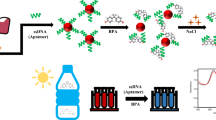
Based on the inhibition of cetyltrimethyal ammonium ion and the enhancement of RNA-aptamer, the differences of the peroxidase-like activities of AuNPs can be greatly enlarged with and without MG, by which a colorimetric aptasensor can be constructed for the detection of Malachite Green (MG).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Malachite green (MG), extensively used in aquaculture as antiparasitic, antifungal and antibacterial agents, has potential carcinogenic, genotoxic, mutagenic and teratogenic properties on human health [1]. Its wide toxicity spectrum may affect aquatic living organisms, causing some organ and tissue damages, reproduction and growth abnormalities [2, 3]. For this reason, MG has been banned in aquaculture in several countries including China. Many analytical methods, including high-performance liquid chromatography (HPLC) [4], HPLC-mass spectrometry (HPLC-MS) [5] and gas chromatography tandem mass spectrometry (GC-MS) [6], have routinely been used for the detection of MG with high sensitivity and selectivity. ELISA is a sensitive and selective method that has been used for rapid screening of MG using antibody as a recognition element [7], but the instable biological antibody and harsh test conditions may limit the applications of ELISA method.
AuNPs, with versatile properties, have been widely used in biosensing [8], biological imaging [9], and catalysis [10]. Particularly, the activities of enzyme mimics of AuNPs have attracted great attention in biochemical studies [11]. For example, citrate-capped AuNPs is used as a mimetic peroxidase which can catalyz the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) by H2O2 to yield a blue color in aqueous solution [12, 13].
Aptamers are DNA/RNA molecules selected from random-sequence nucleic acid libraries [14]. These aptamers have the abilities to recognize their cognate targets with high affinity and specificity [15]. Aptamers are found to greatly enhance the peroxidase-like activity of AuNPs, which have been used to construct various aptasensors [12, 16]. However, our previous study found that the enhancement is not as high as expected, leading to a slight difference in solution color for TMB oxidation.
In the present work, a cationic surfactant specifically by hexadecyl trimethyl ammonium bromide (CTAB) is introduced to enlarge the detection signals of aptasensor. By hexadecyl trimethyl ammonium ion, CTAB can induce the aggregation of negatively charged AuNPs, leading to the inhibition of the catalytic activity of AuNPs. However, the catalytic activity of AuNPs may be enhanced after the addition of RNA-aptamer against MG, because the negatively charged RNA-aptamer binds to CTAB to prevent the aggregation of AuNPs, which results in a dark blue in the catalytic reaction of TMB by H2O2. In the presence of MG, RNA-aptamer preferentially and specifically binds to MG, leading to the inhibition of catalytic activity of AuNPs with a light blue color in solution. To validate the concept, a colorimetric aptasensor is constructed and has been applied to the detection of MG in aquaculture water.
Experimental
Materials and reagents
RNA-aptamer against MG with a sequence of 5′- UCCCG ACUGG AACAG GUAAC GAAUG GA -3′ was purchased from Sangon Biotechnology Co., Ltd., China (https://www.sangon.com). TMB was bought from Beijing lark technology Co., Ltd., China (https://jkchemical.cn.gongchang.com). H2O2 and acetic acid were purchased from Xilong Scientific Co., Ltd., China (http://www.xlhg.com). MG, tetrachloroauric (III) acid tetrahydrate (HAuCl4), trisodium citrate dihydrate and diethyl pyrocarbonate (DEPC) were purchased from Aladdin Reagent Co., Ltd., China (http://www.labgogo.com/News/new_1278.html). Sodium acetate anhydrous (NaAc) was purchased from Shanghai chemical reagent factory, China (http://1062031.co.sonhoo.com/company_web/index-1062032.html). CTAB was obtained from Sinopharm group chemical reagent Co. Ltd., China (https://www.reagent.com.cn). All reagents were of analytical grade without further purification. DEPC water (1‰, w/v) was used for solution preparation and was then sterilized with high pressure. 96-well polystyrene microplates were purchased from Xiamen exhibition chemical Co., Ltd., China (http://liudeqi2006.cn.gtobal.com). All glass and plastic products were soaked with DEPC water overnight and dried for later use.
Instruments
The UV-Vis absorption spectra were performed on a SpectraMax plus 384 microplate reader (Molecular Devices, Silicon valley, USA). The MS7-H550-PRO Magnetic stirrer (Dragon xingchuang experiment instrument Co., Ltd., Beijing, China) was used for the preparation of AuNPs. The centrifuge used was a 5417R Desktop high-speed freezing centrifuge (Cence, Changsha, China). Transmission electron microscopy (TEM) images were recorded with a JEM-2100 micro-scope (Japan Electron Optics Laboratory, Japan). pH values of the buffers were adjusted using a FE 28 pH meter (Mettler Toledo, Switzerland).
Preparation of citrate-capped gold nanoparticles (AuNPs)
Citrate-capped AuNPs was prepared according to a method [17] with some modification. Briefly, 10 mL of 38.8 mmol L−1 sodium citrate was mixed with 100 mL of 1 mmol L−1 HAuCl4 at boiling state. The mixture was continuously heated and stirred until the solution turned to a color of wine-red. After cooling to room temperature, AuNPs was obtained. The solution of AuNPs was stored at 4 °C before use. The concentration of the prepared AuNPs was estimated to be 14.0 nmol L−1 based on an extinction coefficient of 2.7 × 108 mol−1 cm−1 [18].
Analytical procedures
20 μL of MG solutions (prepared with 10 mmol L−1 NaAc-HAc, pH 6.5) at different concentrations was mixed with 30 μL of RNA-aptamer (0.1 μmol L−1) in a microplate for 15 min. Subsequently, 20 μL of AuNPs (10 nmol L−1) was added. After incubated for 15 min, 10 μL of CTAB (70 μmol L−1) was added, and the mixture was fully stirred for 15 min. Finally, 60 μL of TMB (2.0 mmol L−1) and 60 μL of H2O2 (2.0 mol L−1) prepared with 10 mmol L−1 NaAc-HAc (pH 4.0) were added in order sequence. After reaction for 20 min, the solution was measured with a microplate reader. The absorbance at 650 nm was read. All the reactions were carried out at room temperature.
Real samples pretreatment and detection
Two kinds of aquaculture water samples including fresh water and seawater were collected from local fish farming ponds. The samples were filtered through 0.45 μm nylon membranes before use. For recovery tests, 1 mL of aquaculture water samples was spiked with MG at three levels (0.05, 0.10 and 0.50 μmol L−1). 20 μL of sample solutions was measured using the analytical procedures as described above.
Results and discussion
Principle of Malachite Green (MG) detection
The sensing mechanism is illustrated as shown in Fig. 1. TMB, a commonly used peroxidase substrate, can be oxidized by H2O2 through many peroxidases as catalysts including biological enzymes and mimic enzymes. The TMB oxidation produces a color change from colorless to blue with a new absorbance at 650 nm [19]. With intrinsic peroxidase-like activity, AuNPs can be used as a mimic enzyme to effectively catalyze the oxidation reaction of TMB by H2O2 [20], generating various colorimetric sensors in many applications [12, 19, 21]. In addition, the peroxidase-like activity of AuNPs can be enhanced by DNA/RNA-based aptamers, which may be favorable for the catalysis in the oxidation reaction [22, 23]. However, the enhancement of the peroxidase-like activity of AuNPs by aptamer is not as obvious as expected based on our experiment. Similar blue color solution of TMB and H2O2 reaction system was observed in practice. In order to expand the differences, CTAB is introduced to inhibit the peroxidase-like activity of AuNPs as shown in Fig. 1. Zeta potentials in Table 1 indicate that the positively charged CTAB can be adsorbed on the surface of citrate-capped AuNPs which inhibits the peroxidase-like activity of AuNPs. However, the peroxidase-like activity of AuNPs is greatly enhanced after the addition of RNA-aptamer because of the binding of cetyltrimethyal ammonium ions with negatively charged RNA-aptamer through electrostatic attraction, producing a dark blue color in the oxidation reaction of TMB and H2O2. In the presence of MG, RNA-aptamer can specifically bind to MG to form a “duplex structures” [24], leading to the inhibition of enzymatic activity of AuNPs again. The concentrations of MG can be quantified through the absorbance at 650 nm or through the color of solutions.
Absorbance characteristics
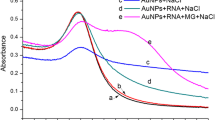
To confirm the feasibility of our concept, the spectral responses of AuNPs at different conditions are shown in Fig. 2. With the characteristic of strong surface plasma resonance, the prepared AuNPs have an absorption peak at 520 nm with a red color as shown in Fig. 2a. After the addition of CTAB, the color changes from red to blue and the absorption peak of AuNPs consequently shifts from 520 nm to 740 nm, which is attributed to the aggregation of AuNPs. With the addition of RNA-aptamer, the red color recovers, implying that AuNPs disperse again in the solution. However, in the presence of MG, the color changes from red to blue again because of the specific binding of RNA-aptamer with MG and the aggregation of AuNPs triggered by the released cetyltrimethyal ammonium ions.
TEM images of AuNPs in Fig. 3 show that the citrate-capped AuNPs with average diameter of 13 nm disperse well in solutions as shown in Fig. 3a, but the particles completely aggregate after mixed with CTAB (Fig. 3b). When the RNA-aptamer is introduced, the AuNPs re-disperse (Fig. 3c). However, in the presence of MG, the AuNPs aggregate again as shown in Fig. 3d.
For further confirming the sensing feasibility, AuNPs as a mimic enzyme are tested as shown in Fig. 2b. AuNPs can catalyze the oxidation reaction of TMB and H2O2 to develop a blue color in solution with a maximum absorbance at 650 nm. After mixed with CTAB, AuNPs loses its mimic enzyme activity. But the catalytic activity of AuNPs can be greatly enhanced with an absorbance at 650 nm similar to that of the binary solution of AuNPs and RNA-aptamer when RNA-aptamer is added. The results confirm the presence of interaction between CTAB and RNA-aptamer. The results also confirm that the released AuNPs may present higher catalytic activity compared with that of AuNPs alone because of the enhance effect by RNA-aptamer. After the addition of MG, the absorbance at 650 nm decreases which is due to the specific binding of MG to RNA-aptamer, resulting in the re-aggregation of AuNPs. Therefore, MG can be detected with the colorimetric aptasensor.
Optimization of detection conditions
△A650 (A0-AMG, where A0 and AMG were the absorbance at 650 nm in the absence and presence of MG, respectively) was used as indexes to optimize the detection conditions, including the concentrations of RNA-aptamer, AuNPs and CTAB, and the amounts of TMB and H2O2. Respective data and figures are given in the Electronic Supporting Material. The following experimental conditions gave the best results: (a) 0.1 μmol L−1 of RNA-aptamer; (b) 70 μmol L−1 of CTAB and 10 nmol L−1 of AuNPs; (c) 2 mol L−1 of H2O2 and 2 mmol L−1 of TMB.
Calibration plot and detection limit
Under the optimal conditions, the colorimetric aptasensor was used to detect MG by measuring the absorbance at 650 nm. Figure 4a shows the absorption spectra with MG at different concentrations, in which the absorbance at 650 nm decreases with MG concentrations ranging from 0.01 to 0.5 μmol L−1. A linear equation of △A650 = 0.43C + 0.03 (R2 = 0.998) is constructed as shown in Fig. 4b. The limit of detection (LOD) is 1.8 nmol L−1 estimated with 3σ/k (where σ is the standard deviation of nine parallel blank measurements with MG concentration of 10 nmol L−1 and k is the slope of the linear equation). The LOD is better than that of the fluorescence method and colorimetric biosensor, and even better than that of GC-MS. In addition, the LOD is close to those of HPLC, LC-MS, and commercial ELISA as shown in Table 2, but the detection time was far less than that of the ELISA method. Therefore, the method of colorimetric aptasensor is sensitive and rapid for MG detection.
Selectivity
Some chemicals including sulfaguanidine (SG), sulfanilamide (SAs), oxytetracycline hydrochloride (OTH) and chloramphenicol (CAP), which are legally or illegally used in fish farming, were chosen for selectivity tests. Results in Fig. 5 show that the absorbance for MG is much lower compared with those of the four chemicals, and the coexistence of the four chemicals does not influence on the MG detection. In addition, the possible influences of cations on MG detection were tested. Results in Table 3 show that more than 4800 times of Na+, K+, Ca2+, Mg2+ and Ba2+ do not interfere with MG detection, but Fe3+ (≥0.4 mmol L −1) may have an interference because of its enzyme mimics activity [27]. Therefore, the colorimetric aptasensor is specific for MG detection which may be attributed to the specific binding of RNA-aptamer to MG.
Aquaculture analysis
To evaluate the practical applicability, two kinds of aquaculture water samples including fresh water and seawater (physicochemical parameters were shown in Table S1) were spiked with MG at three levels (0.05, 0.1, 0.5 μmol L−1), and MG were respectively measured with the as-prepared colorimetric aptasensor, HPLC [28] and ELISA. As shown in Table 4, the recoveries range from 88% to 135% with RSDs<5.0% for the method of colorimetric aptasensor, which is in agreement with those of HPLC and ELISA (for fresh water samples). It is noted that the detection of MG in seawater sample by a commercial ELISA kit is infeasible as shown in Table 4, which may be due to the interference of seawater matrix. In order to test the repeatability, the colorimetric aptasensor were carried out in intraday and interday assays with spiked water samples, results in Table 5 show that RSDs are less than 3.0% for both intraday and interday assays, demonstrating that the colorimetric aptasensor is accurate with excellent repeatability. However, the colorimetric aptasensor can’t be used to detect MG in fish samples based on our experiments. It is probably due to the presence of complicated components in fish, especially various RNA enzymes that will decompose RNA-aptamer may be the main reasons.
Conclusions
A colorimetric aptasensor based on CTAB-inhibited peroxidase-like catalysis of AuNPs was constructed for the detection of MG. Based on the inhibition by cetyltrimethyal ammonium ion and the enhancement by RNA-aptamer the differences of the peroxidase-like activities of AuNPs can be greatly enlarged with and without MG. Based on the colorimetric aptasensor, MG in aquaculture water can be quantified specifically and accurately.
References
Xiao D, Jiang Y, Bi Y (2018) Molecularly imprinted polymers for the detection of illegal drugs and additives: a review. Microchim Acta 185:247
Bergwerff AA, Scherpenisse P (2003) Determination of residues of malachite green in aquatic animals. J Chromatogr B 788:351–359
Srivastava S, Sinha R, Roy D (2004) Toxicological effects of malachite green. Aquat Toxicol 66:319–329
Martínez Bueno MJ, Herrera S, Uclés A, Agüera A, Hernando MD, Shimelis O, Rudolfsson M, Fernández-Alba AR (2010) Determination of malachite green residues in fish using molecularly imprinted solid-phase extraction followed by liquid chromatography-linear ion trap mass spectrometry. Anal Chim Acta 665:47–54
Tarbin J, Barnes K, Bygrave J (1998) Screening and confirmation of triphenylmethane dyes and their leuco metabolites in trout muscle using HPLC-vis and ESP-LC-MS. Analyst 123:2567–2571
Aprea C, Colosio C, Mammone T, Minoia C, Maroni M (2002) Biological monitoring of pesticide exposure: a review of analytical methods. J Chromatogr B 769:191–219
Li L, Peng A, Lin Z, Zhong H, Chen X, Huang Z (2017) Biomimetic ELISA detection of malachite green based on molecularly imprinted polymer film. Food Chem 229:403–408
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277:1078–1081
Jain PK, Lee KS, El-Sayed IH, El-Sayed MA (2006) Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J Phys Chem B 110:7238–7248
Khan MM, Lee J, Cho MH (2013) Electrochemically active biofilm mediated bio-hydrogen production catalyzed by positively charged gold nanoparticles. Int J Hydrogen Energ 38:5243–5250
Lin Y, Ren J, Qu X (2014) Nano-gold as artificial enzymes: hidden talents. Adv Mater 26:4200–4217
Hu J, Ni P, Dai H, Sun Y, Wang Y, Jiang S, Li Z (2015) Aptamer-based colorimetric biosensing of abrin using catalytic gold nanoparticles. Analyst 140:3581–3586
Wang C, Chen D, Wang Q, Tan R (2017) Kanamycin detection based on the catalytic ability enhancement of gold nanoparticles. Biosens Bioelectron 91:262–267
Hu JT, Dai HC, Sun YJ, Ni PJ, Wang YL, Jiang S, Li Z (2014) Highly sensitive and rapid visual detection of ricin using unmodified gold nanoparticle probes. RSC Adv 4:43998– 44003
Wu C, Han D, Chen T, Peng L, Zhu G, You M, Qiu L, Sefah K, Zhang X, Tan W (2013) Building a multifunctional Aptamer-based DNA Nanoassembly for targeted cancer therapy. J Am Chem Soc 135:18644–18650
Liu B, Liu J (2015) Accelerating peroxidase mimicking nanozymes using DNA. Nanoscale 7:13831–13835
Lubin AA, Plaxco KW (2010) Folding-based electrochemical biosensors: the case for responsive nucleic acid architectures. Acc Chem Res 43:496–505
Maye MM, Han L, Kariuki NN, Ly NK, Chan W, Luo J, Zhong C (2003) Gold and alloy nanoparticles in solution and thin film assembly: spectrophotometric determination of molar absorptivity. Anal Chim Acta 496:17–27
Hu J, Ni P, Dai H (2015) A facile label-free colorimetric aptasensor for ricin based on the peroxidase-like activity of gold nanoparticles. RSC Adv 5:11541–11548
Astruc D, Lu F, Aranzaes JR (2005) Nanoparticles as recyclable catalysts: the frontier between homogeneous and heterogeneous catalysis. Angew Chem Int Ed 44:7852–7872
Wang C, Dong X, Liu Q, Wang K (2015) Label-free colorimetric aptasensor for sensitive detection of ochratoxin a utilizing hybridization chain reaction. Anal Chim Acta 860:83–88
Hizir MS, Top M, Balcioglu M, Rana M, Robertson NM, Shen F, Sheng J, Yigit MV (2015) Multiplexed activity of perAuxidase: DNA-capped AuNPs act as adjustable peroxidase. Anal Chem 88:600–605
Yang J, Lu Y, Ao L (2017) Colorimetric sensor array for proteins discrimination based on the tunable peroxidase-like activity of AuNPs-DNA conjugates. Sensor Actuat B-Chem 245:66–73
Le Thao Nguyen N, Park CY, Park JP (2018) Synergistic molecular assembly of an aptamer and surfactant on gold nanoparticles for the colorimetric detection of trace levels of As3+ ions in real samples. New J Chem 42:11530–11538
Yan Jun J, Li N, Liu SG, Han L, Xiao N, Luo HQ, Li NB (2018) Ratiometric fluorescence method for malachite green detection based on dual-emission BSA-protected gold nanoclusters. Sensors Actuators B Chem 275:244–250
Jia J, Yan S, Lai X, Xu Y, Liu T, Xiang Y (2018) Colorimetric Aptasensor for Detection of Malachite Green in Fish Sample Based on RNA and Gold Nanoparticles. Food Anal Methods 11(6):1668–1676
Attar F, Shahpar MG, Rasti B (2019) Nanozymes with intrinsic peroxidase-like activities. J Mol Liq 278:130–144
Li L, Lin Z, Chen X, Zhang H, Lin Y, Lai Z, Huang Z (2015) Molecularly imprinted polymers for extraction of malachite green from fish samples prior to its determination by HPLC. Microchim Acta 182:1791–1796.
Acknowledgements
This research was supported by the Foundation from the Science and Technology Planning Project of Fujian Province, China (2016Y0064), the Natural Science Foundation of Fujian Province of China (2018 J01432, 2017 J01633), National Key R and D Program of China (2018YFD0901003), the Science and Technology Planning Project of Xiamen, China (3502Z20183031), and the National Undergraduate Training Programs for Innovation and Entrepreneurship (201610390042, 201710390022, 201810390071, 20181xj008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 206 kb)
Rights and permissions
About this article
Cite this article
Zhao, C., Hong, Cy., Lin, Zz. et al. Detection of Malachite Green using a colorimetric aptasensor based on the inhibition of the peroxidase-like activity of gold nanoparticles by cetyltrimethylammonium ions. Microchim Acta 186, 322 (2019). https://doi.org/10.1007/s00604-019-3436-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3436-3